Use generative adversarial networks (GANs) to generate real-valued time series. The GAN uses RNNs for both encoder and decoder (specifically LSTMs).
Primary dependencies: tensorflow, scipy, numpy, pandas. See requirements.txt for specific versions.
This code is tested on both Python 2.7.16 and Python 3.6.6.
Simplest route to running code (Linux/Mac):
git clone git@github.com:ratschlab/RGAN.git
cd RGAN
mkdir experiments/parameters experiments/data
python experiment.py --settings_file sine
Here, experiments/settings/sine.txt is the settings file for generating sine waves.
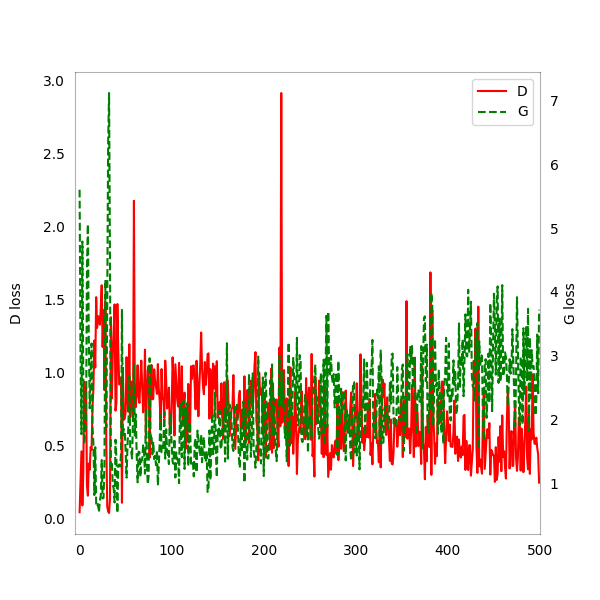
Evolution of Discriminator (D) and Generator (G ) training loss:
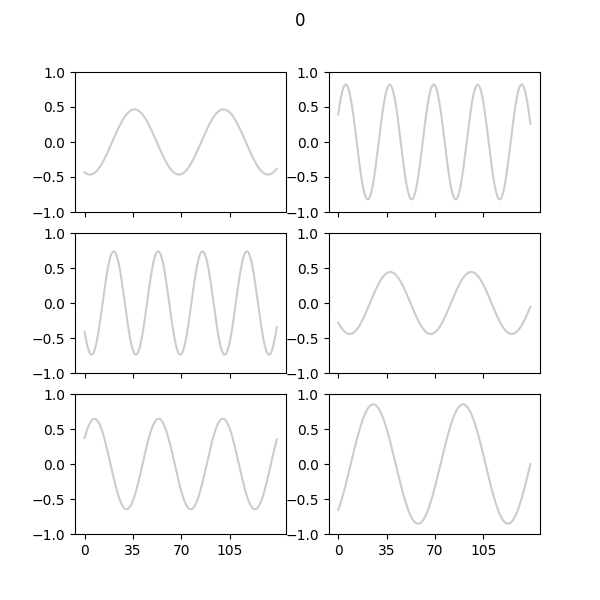
Random samples of synthetic sine waves generated at each epoch:
Random sample of real sine waves:
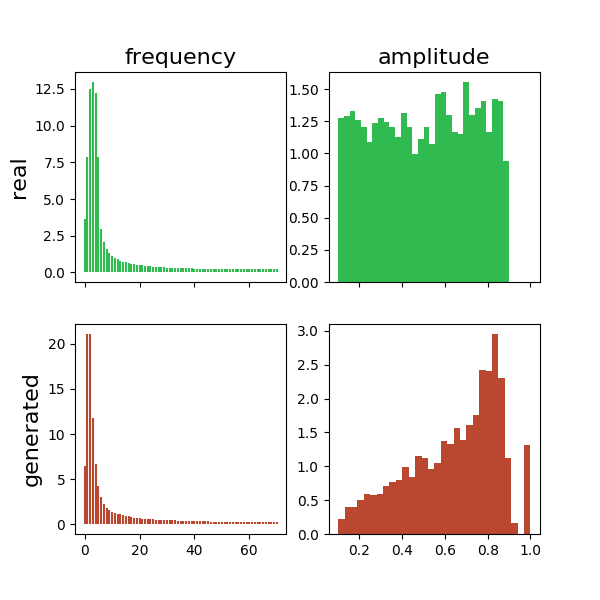
Frequency and amplitude distributions of real and generated sine waves:
.csv files of real and generated sine waves saved to experiments/data.
Get MNIST as CSVs here: https://pjreddie.com/projects/mnist-in-csv/
python experiment.py --settings_file mnistfull
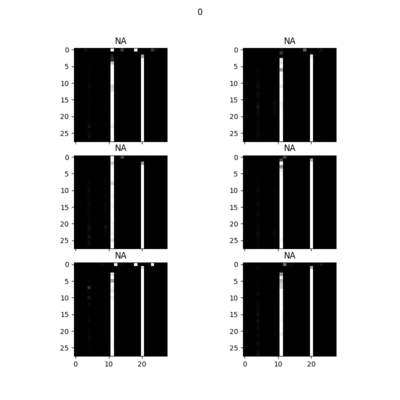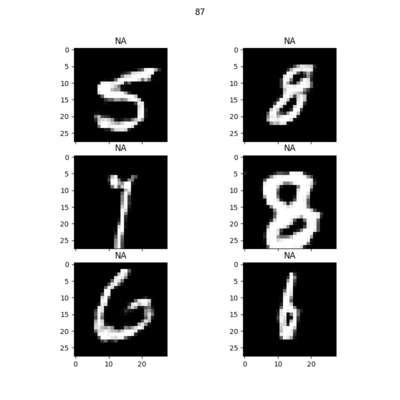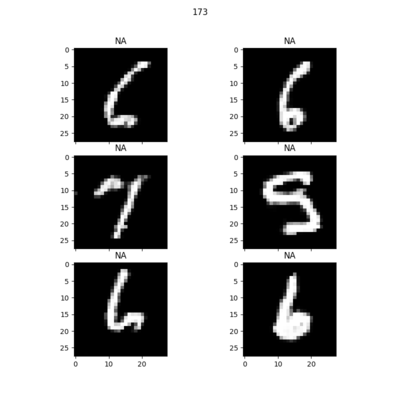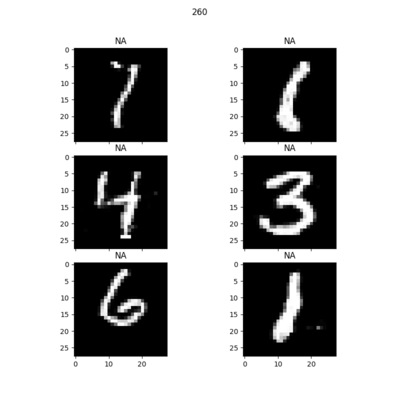
Random samples of synthetic MNIST digits generated at each epoch:
The main script is experiment.py - this parses many options, loads and preprocesses data as needed, trains a model, and does evaluation. It does this by calling on some helper scripts:
data_utils.py: utilities pertaining to data: generating toy data (e.g. sine waves, GP samples), loading MNIST and eICU data, doing test/train split, normalising data, generating synthetic data to use in TSTR experimentsmodel.py: functions for defining ML models, i.e. the tensorflow meat, defines the generator and discriminator, the update steps, and functions for sampling from the model and 'inverting' points to find their latent-space representationsplotting.py: visualisation scripts using matplotlibmmd.py: for maximum-mean discrepancy calculations, mostly taken from https://github.com/dougalsutherland/opt-mmd
Other scripts in the repo:
eval.py: functions for evaluating the RGAN/generated data, like testing if the RGAN has memorised the training data, comparing two models, getting reconstruction errors, and generating data for visualistions of things like varying the latent dimensions, interpolating between input samplesmod_core_rnn_cell_impl.py: this is a modification of the same script from TensorFlow, modified to allow us to initialise the bias in the LSTM (required for saving/loading models)kernel.py: some playing around with kernels on time seriestf_ops.py: required byeugenium_mmd.py
This repository is forked from ratschlab/RGAN the repo for the paper, Real-valued (Medical) Time Series Generation with Recurrent Conditional GANs_, by Stephanie L. Hyland* (@corcra), Cristóbal Esteban* (@cresteban), and Gunnar Rätsch (@ratsch), from the Ratschlab, also known as the Biomedical Informatics Group at ETH Zurich.