📜 arXiv 📦 Python package 📚 RTDL (other projects on tabular DL)
This is the official implementation of the paper "Revisiting Deep Learning Models for Tabular Data".
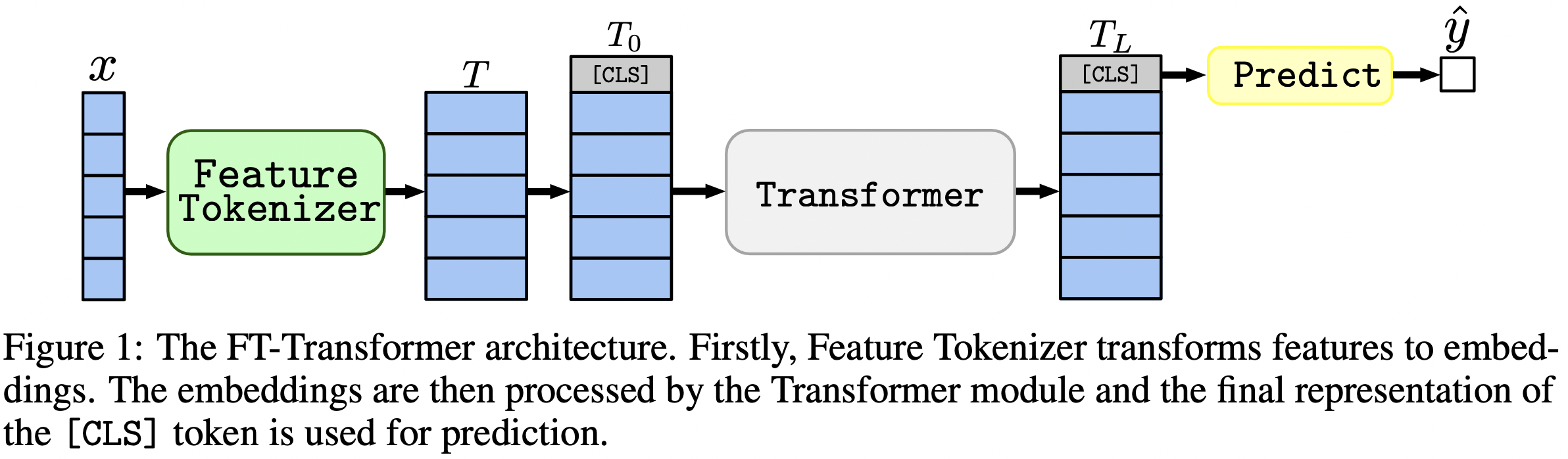
In one sentence: MLP-like models are still good baselines, and FT-Transformer is a new powerful adaptation of the Transformer architecture for tabular data problems.
The paper focuses on architectures for tabular data problems. The results:
- A simple tuned MLP is still a good baseline: it performs on par with or even better than most of sophisticated architectures.
- ResNet (an MLP-like model with skip connections and batch normalizations) further highlights this point: MLP-like models are good baselines for tabular deep learning, and prior work does not outperform them.
- FT-Transformer is a new architecture which changes this status quo:
- on benchmarks, it demonstrated the best average performance among deep models (including the aforementioned MLP-like baselines);
- on the datasets where GBDT (gradient-booosted decision trees) dominates over DL models, FT-Transformer reduces (not completely) the gap between GBDT and DL.
- FT-Transformer is slower than MLP-like models
The Python package in the package/ directory
is the recommended way to use the paper in practice and for future work.
The rest of the document:
The output/ directory contains numerious results and (tuned) hyperparameters
for various models and datasets used in the paper.
For example, let's explore the metrics for the MLP model.
First, let's load the reports (the stats.json files):
import json
from pathlib import Path
import pandas as pd
df = pd.json_normalize([
json.loads(x.read_text())
for x in Path('output').glob('*/mlp/tuned/*/stats.json')
])Now, for each dataset, let's compute the test score averaged over all random seeds:
print(df.groupby('dataset')['metrics.test.score'].mean().round(3))The output exactly matches Table 2 from the paper:
dataset
adult 0.852
aloi 0.954
california_housing -0.499
covtype 0.962
epsilon 0.898
helena 0.383
higgs_small 0.723
jannis 0.719
microsoft -0.747
yahoo -0.757
year -8.853
Name: metrics.test.score, dtype: float64
The above approach can also be used to explore hyperparameters to get intuition on typical hyperparameter values for different algorithms. For example, this is how one can compute the median tuned learning rate for the MLP model:
Note
For some algorithms (e.g. MLP), more recent projects offer more results that can be explored in a similar way. For example, see this paper on TabR.
Warning
Use this approach with caution. When studying hyperparameter values:
- Beware of outliers.
- Take a look at raw unaggregated values to get intuition on typical values.
- For a high-level overview, plot the distribution and/or compute multiple quantiles.
print(df[df['config.seed'] == 0]['config.training.lr'].quantile(0.5))
# Output: 0.0002161505605899536Note
This section is long. Use the "Outline" feature on GitHub on in your text editor to get an overview of this section.
The code is organized as follows:
bin:- training code for all the models
ensemble.pyperforms ensemblingtune.pyperforms hyperparameter tuning- code for the section "When FT-Transformer is better than ResNet?" of the paper:
analysis_gbdt_vs_nn.pyruns the experimentscreate_synthetic_data_plots.pybuilds plots
libcontains common tools used by programs inbinoutputcontains configuration files (inputs for programs inbin) and results (metrics, tuned configurations, etc.)packagecontains the Python package for this paper
Install conda
export PROJECT_DIR=<ABSOLUTE path to the repository root>
# example: export PROJECT_DIR=/home/myusername/repositories/revisiting-models
git clone https://github.com/yandex-research/tabular-dl-revisiting-models $PROJECT_DIR
cd $PROJECT_DIR
conda create -n revisiting-models python=3.8.8
conda activate revisiting-models
conda install pytorch==1.7.1 torchvision==0.8.2 cudatoolkit=10.1.243 numpy=1.19.2 -c pytorch -y
conda install cudnn=7.6.5 -c anaconda -y
pip install -r requirements.txt
conda install nodejs -y
jupyter labextension install @jupyter-widgets/jupyterlab-manager
# if the following commands do not succeed, update conda
conda env config vars set PYTHONPATH=${PYTHONPATH}:${PROJECT_DIR}
conda env config vars set PROJECT_DIR=${PROJECT_DIR}
conda env config vars set LD_LIBRARY_PATH=${CONDA_PREFIX}/lib:${LD_LIBRARY_PATH}
conda env config vars set CUDA_HOME=${CONDA_PREFIX}
conda env config vars set CUDA_ROOT=${CONDA_PREFIX}
conda deactivate
conda activate revisiting-modelsThis environment is needed only for experimenting with TabNet. For all other cases use the PyTorch environment.
The instructions are the same as for the PyTorch environment (including installation of PyTorch!), but:
python=3.7.10cudatoolkit=10.0- right before
pip install -r requirements.txtdo the following:pip install tensorflow-gpu==1.14- comment out
tensorboardinrequirements.txt
LICENSE: by downloading our dataset you accept licenses of all its components. We do not impose any new restrictions in addition to those licenses. You can find the list of sources in the section "References" of our paper.
- Download the data:
wget https://www.dropbox.com/s/o53umyg6mn3zhxy/data.tar.gz?dl=1 -O revisiting_models_data.tar.gz - Move the archive to the root of the repository:
mv revisiting_models_data.tar.gz $PROJECT_DIR - Go to the root of the repository:
cd $PROJECT_DIR - Unpack the archive:
tar -xvf revisiting_models_data.tar.gz
This section only provides specific commands with few comments. After completing the tutorial, we recommend checking the next section for better understanding of how to work with the repository. It will also help to better understand the tutorial.
In this tutorial, we will reproduce the results for MLP on the California Housing dataset. We will cover:
- tuning
- evaluation
- ensembling
- comparing models with each other
Note that the chances to get exactly the same results are rather low, however, they should not differ much from ours. Before running anything, go to the root of the repository and explicitly set CUDA_VISIBLE_DEVICES (if you plan to use GPU):
cd $PROJECT_DIR
export CUDA_VISIBLE_DEVICES=0Before we start, let's check that the environment is configured successfully. The following commands should train one MLP on the California Housing dataset:
mkdir draft
cp output/california_housing/mlp/tuned/0.toml draft/check_environment.toml
python bin/mlp.py draft/check_environment.tomlThe result should be in the directory draft/check_environment. For now, the content of the result is not important.
Our config for tuning MLP on the California Housing dataset is located at output/california_housing/mlp/tuning/0.toml.
In order to reproduce the tuning, copy our config and run your tuning:
# you can choose any other name instead of "reproduced.toml"; it is better to keep this
# name while completing the tutorial
cp output/california_housing/mlp/tuning/0.toml output/california_housing/mlp/tuning/reproduced.toml
# let's reduce the number of tuning iterations to make tuning fast (and ineffective)
python -c "
from pathlib import Path
p = Path('output/california_housing/mlp/tuning/reproduced.toml')
p.write_text(p.read_text().replace('n_trials = 100', 'n_trials = 5'))
"
python bin/tune.py output/california_housing/mlp/tuning/reproduced.tomlThe result of your tuning will be located at output/california_housing/mlp/tuning/reproduced, you can compare it with ours: output/california_housing/mlp/tuning/0. The file best.toml contains the best configuration that we will evaluate in the next section.
Now we have to evaluate the tuned configuration with 15 different random seeds.
# create a directory for evaluation
mkdir -p output/california_housing/mlp/tuned_reproduced
# clone the best config from the tuning stage with 15 different random seeds
python -c "
for seed in range(15):
open(f'output/california_housing/mlp/tuned_reproduced/{seed}.toml', 'w').write(
open('output/california_housing/mlp/tuning/reproduced/best.toml').read().replace('seed = 0', f'seed = {seed}')
)
"
# train MLP with all 15 configs
for seed in {0..14}
do
python bin/mlp.py output/california_housing/mlp/tuned_reproduced/${seed}.toml
doneOur directory with evaluation results is located right next to yours, namely, at output/california_housing/mlp/tuned.
# just run this single command
python bin/ensemble.py mlp output/california_housing/mlp/tuned_reproducedYour results will be located at output/california_housing/mlp/tuned_reproduced_ensemble, you can compare it with ours: output/california_housing/mlp/tuned_ensemble.
Use the approach described here
to summarize the results of the conducted experiment
(modify the path filter in .glob(...) accordingly: tuned -> tuned_reproduced).
Similar steps can be performed for all models and datasets. The tuning process is
slightly different in the case of grid search: you have to run all desired
configurations and manually choose the best one based on the validation performance.
For example, see output/epsilon/ft_transformer.
You should run Python scripts from the root of the repository. Most programs expect a
configuration file as the only argument. The output will be a directory with the same
name as the config, but without the extention. Configs are written in
TOML. The lists of possible arguments for the programs are not
provided and should be inferred from scripts (usually, the config is represented with
the args variable in scripts). If you want to use CUDA, you must explicitly set the
CUDA_VISIBLE_DEVICES environment variable. For example:
# The result will be at "path/to/my_experiment"
CUDA_VISIBLE_DEVICES=0 python bin/mlp.py path/to/my_experiment.toml
# The following example will run WITHOUT CUDA
python bin/mlp.py path/to/my_experiment.tomlIf you are going to use CUDA all the time, you can save the environment variable in the Conda environment:
conda env config vars set CUDA_VISIBLE_DEVICES="0"The -f (--force) option will remove the existing results and run the script from scratch:
python bin/whatever.py path/to/config.toml -f # rewrites path/to/configbin/tune.py supports continuation:
python bin/tune.py path/to/config.toml --continueFor all scripts, stats.json is the most important part of output. The content varies
from program to program. It can contain:
- metrics
- config that was passed to the program
- hardware info
- execution time
- and other information
Predictions for train, validation and test sets are usually also saved.
Now, you know everything you need to reproduce all the results and extend this repository for your needs. The tutorial also should be more clear now. Feel free to open issues and ask questions.
@inproceedings{gorishniy2021revisiting,
title={Revisiting Deep Learning Models for Tabular Data},
author={Yury Gorishniy and Ivan Rubachev and Valentin Khrulkov and Artem Babenko},
booktitle={{NeurIPS}},
year={2021},
}