Hello There! Welcome to GRIEVOUS, your command-line general to order the variant galaxy. Specifically, GRIEVOUS is your command-line tool for guaranteeing cross-dataset variant consistency in genomic datasets.
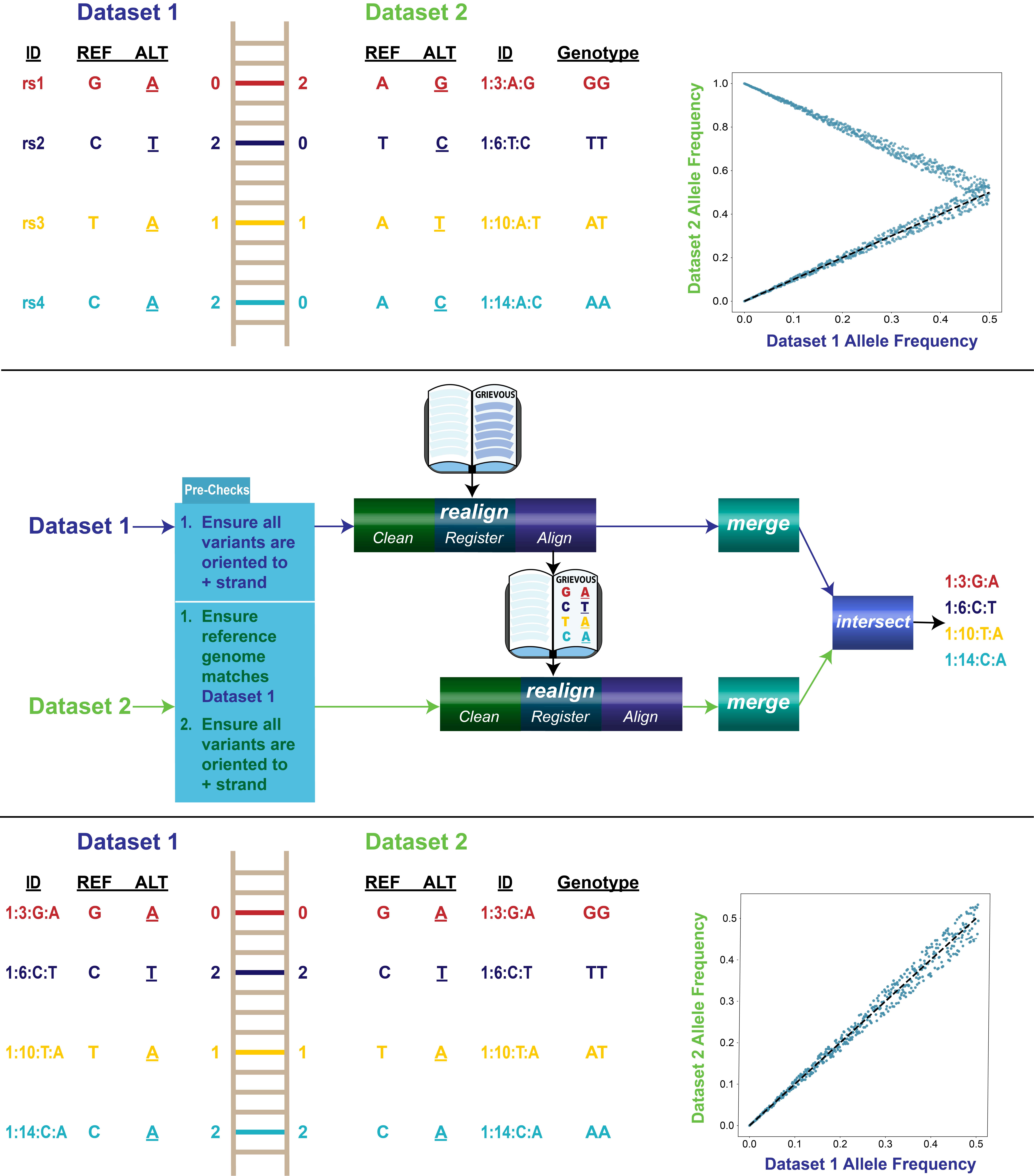
The synchronization of variant indexing and allele assignments is critical for multi-genomic dataset integration. Previously, the burdens of generating unified variant indices, synchronizing allele assignments, and extracting the set of variants common across all datasets of interest, fell upon the researcher. GRIEVOUS was designed to abstract, automate, and ameliorate these tasks. Succinctly, GRIEVOUS homogenizes variant indices and synchronizes biallelic SNP allele assignments across all datasets of interest (regardless of the number) consistently, while also handling common-issues such as duplications or multiple-indexing. GRIEVOUS can also be used to extract the set of biallelic SNPs common across all datasets of interest.
- Cross-dataset consistency: Ensures that variant indices and biallelic allele assignments are consistent across all datasets.
- Data compatibility: Works with both summary statistics and genotype files.
- Extraction: Can extract the set of biallelic SNPs common across all genomic datasets of interest.
For instructions on how to install and use GRIEVOUS, jump on over to our documentation and tutorial.
If you found GRIEVOUS helpful in your research, don't forget to cite our paper: GRIEVOUS: Your command-line general for resolving cross-dataset genotype inconsistencies