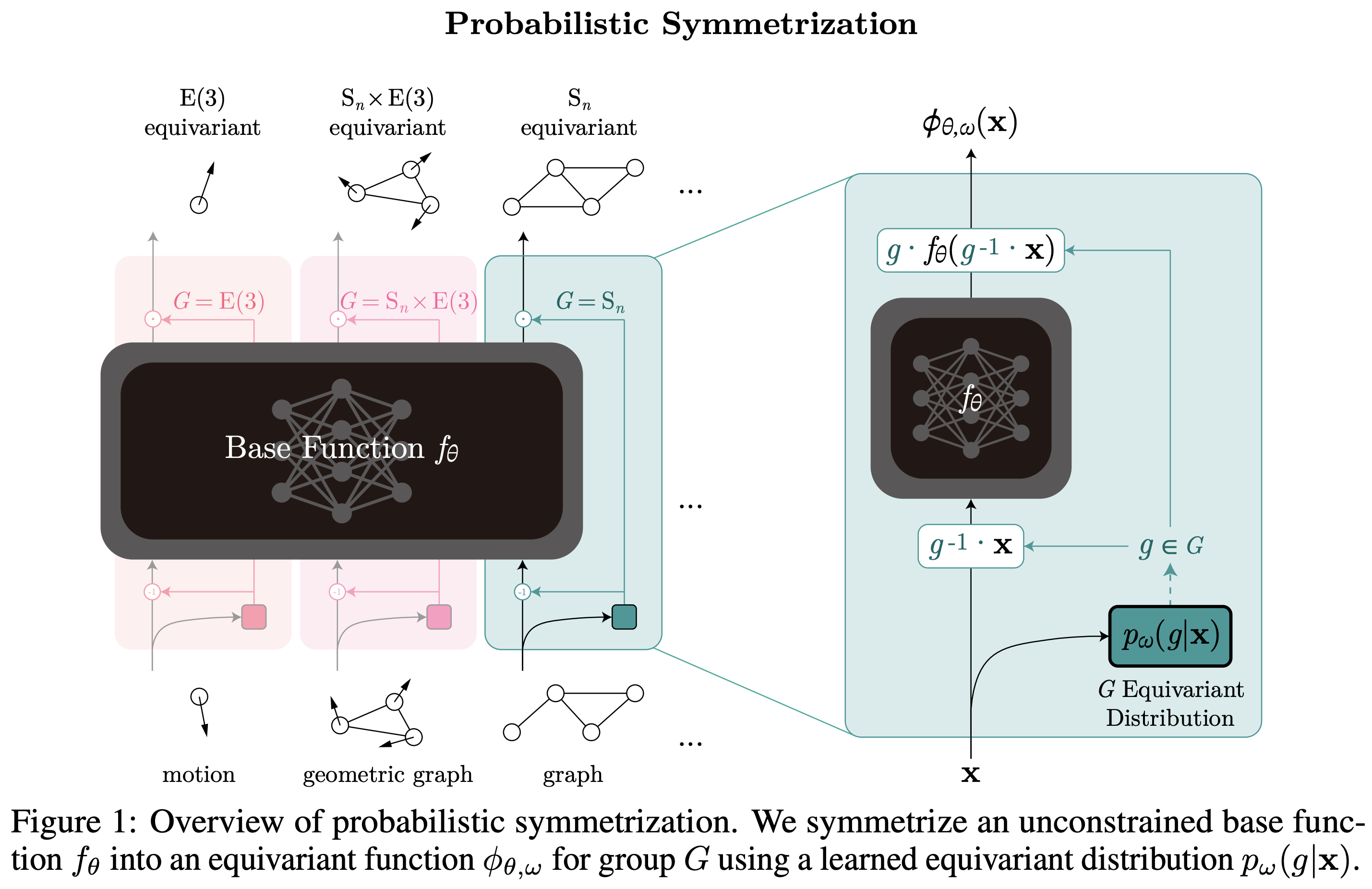
Learning Probabilistic Symmetrization for Architecture Agnostic Equivariance
Jinwoo Kim, Tien Dat Nguyen, Ayhan Suleymanzade, Hyeokjun An, Seunghoon Hong @ KAIST
NeurIPS 2023 (Spotlight Presentation)
Apr 14, 2024
- Fixed evaluation bug for PATTERN and Peptides-func.
- Updated configurations and checkpoints for Peptides-func and Peptides-struct.
Using Docker image (recommended)
docker pull jw9730/lps:latest
docker run -it --gpus all --ipc host --name lps -v /home:/home jw9730/lps:latest bash
# upon completion, you should be at /lps inside the containerUsing Dockerfile
git clone https://github.com/jw9730/lps.git /lps
cd lps
docker build --no-cache --tag lps:latest .
docker run -it --gpus all --ipc host --name lps -v /home:/home lps:latest bash
# upon completion, you should be at /lps inside the containerUsing pip
git clone https://github.com/jw9730/lps.git lps
cd lps
bash install.shGraph isomorphism learning on GRAPH8c, EXP, and EXP-classify (Section 3.1 and Appendix 4.1)
cd scripts
# GRAPH8c
bash graph8c_mlp_canonical.sh
bash graph8c_mlp_ps.sh
bash graph8c_gin_id_canonical.sh
bash graph8c_gin_id_ps.sh
# EXP
bash exp_mlp_canonical.sh
bash exp_mlp_ps.sh
bash exp_gin_id_canonical.sh
bash exp_gin_id_ps.sh
# EXP-classify
bash exp_classify_mlp_canonical.sh
bash exp_classify_mlp_ps_fixed_noise.sh
bash exp_classify_mlp_ps.sh
bash exp_classify_gin_id_canonical.sh
bash exp_classify_gin_id_ps.shParticle dynamics learning on n-body (Section 3.2 and Appendix 4.2)
cd scripts
# n-body
bash nbody_transformer_canonical.sh
bash nbody_transformer_ga.sh
bash nbody_transformer_ps.sh
bash nbody_gnn_canonical.sh
bash nbody_gnn_ga.sh
bash nbody_gnn_ps.shGraph pattern recognition on PATTERN (Section 3.3)
cd scripts
# PATTERN
bash pattern_vit_scratch_ga.sh
bash pattern_vit_scratch_fa.sh
bash pattern_vit_scratch_canonical.sh
bash pattern_vit_scratch_ps.sh
bash pattern_vit_imagenet21k_ga.sh
bash pattern_vit_imagenet21k_fa.sh
bash pattern_vit_imagenet21k_canonical.sh
bash pattern_vit_imagenet21k_ps.shReal-world graph learning on Peptides-func, Peptides-struct, and PCQM-Contact (Section 3.4)
cd scripts
# Peptides-func
bash peptides_func_vit_imagenet21k_ps.sh
# Peptides-struct
bash peptides_struct_vit_imagenet21k_ps.sh
# PCQM-Contact
bash pcqm_contact_vit_imagenet21k_ps.shSupplementary analysis on EXP-classify and inner-symmetric graphs (Appendix 4.3 and 4.4)
cd scripts
# effect of sample size on training and inference
# + additional comparison to group averaging
bash exp_classify_mlp_ps_analysis.sh
# additional comparison to canonicalization
bash automorphism_mlp.shTrained model checkpoints can be found at this link.
To run analysis or testing, please find and download the checkpoints of interest according to the below table.
After that, you can run each experiment script (e.g., bash nbody_transformer_ps.sh) to skip training and run testing.
| Experiment | Download Path |
|---|---|
| n-body | src_synthetic/nbody/experiments/checkpoints/[EXP_NAME]/*.ckpt |
| PATTERN | experiments/checkpoints/gnn_benchmark_pattern/[EXP_NAME]/*.ckpt |
| Peptides-func | experiments/checkpoints/lrgb_peptides_func/[EXP_NAME]/*.ckpt |
| Peptides-struct | experiments/checkpoints/lrgb_peptides_struct/[EXP_NAME]/*.ckpt |
| PCQM-Contact | experiments/checkpoints/lrgb_pcqm_contact/[EXP_NAME]/*.ckpt |
| Supplementary analysis | src_synthetic/graph_separation/experiments/checkpoints/[EXP_NAME]/*.ckpt |
Our implementation uses code from the following repositories:
- Frame Averaging for GRAPH8c, EXP, EXP-classify, and n-body experiments
- Vector Neurons for n-body experiments
- Benchmarking Graph Neural Networks for PATTERN experiments
- Long Range Graph Benchmark for Peptides-func, Peptides-struct, and PCQM-Contact experiments
If you find our work useful, please consider citing it:
@article{kim2023learning,
author = {Jinwoo Kim and Tien Dat Nguyen and Ayhan Suleymanzade and Hyeokjun An and Seunghoon Hong},
title = {Learning Probabilistic Symmetrization for Architecture Agnostic Equivariance},
journal = {arXiv},
volume = {abs/2306.02866},
year = {2023},
url = {https://arxiv.org/abs/2306.02866}
}