Authors: Joe Abbott, Ryan Beck, Hang Hu, Yang Liu, Lixin Lu.
SPEEDCOM is an open source python package that aims to predict the fluorescence emission and absorption spectra of small conjugated organic molecules. These features are predicted using a convolutional neural network, implemented with keras, and trained on data from the PhotochemCAD database. The software has a graphical-user-interface (GUI) where users can input the SMILES string for a given molecule and be returned its predicted spectra and associated characteristic quantities. For further details on the background science, and the operations of our program, please see our use cases.
- Input vaild SMILES of small organic molecules as input for prediction
- Predictions on the maximum absorption/emission peak
- Wrapper functions for a numerial encoding of SMILES and common descriptors generation
- GUI on local host
- Sanitize SMILES input; add alternative input options
- Predictions on multiple peaks of absorption/emission spectra
- Customization of model training with user input data
- Pipelining predictions to facilitate fluorophore design
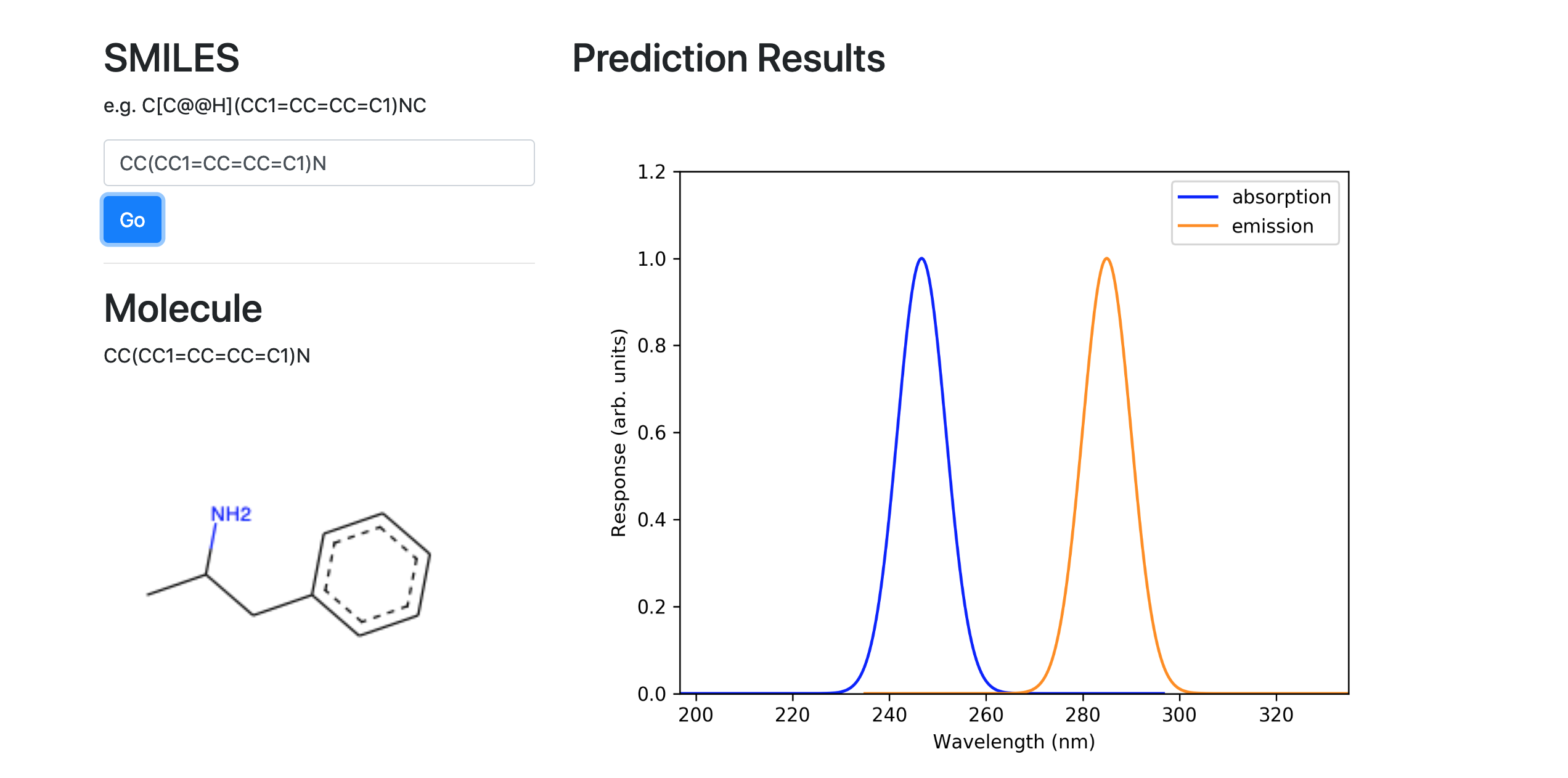
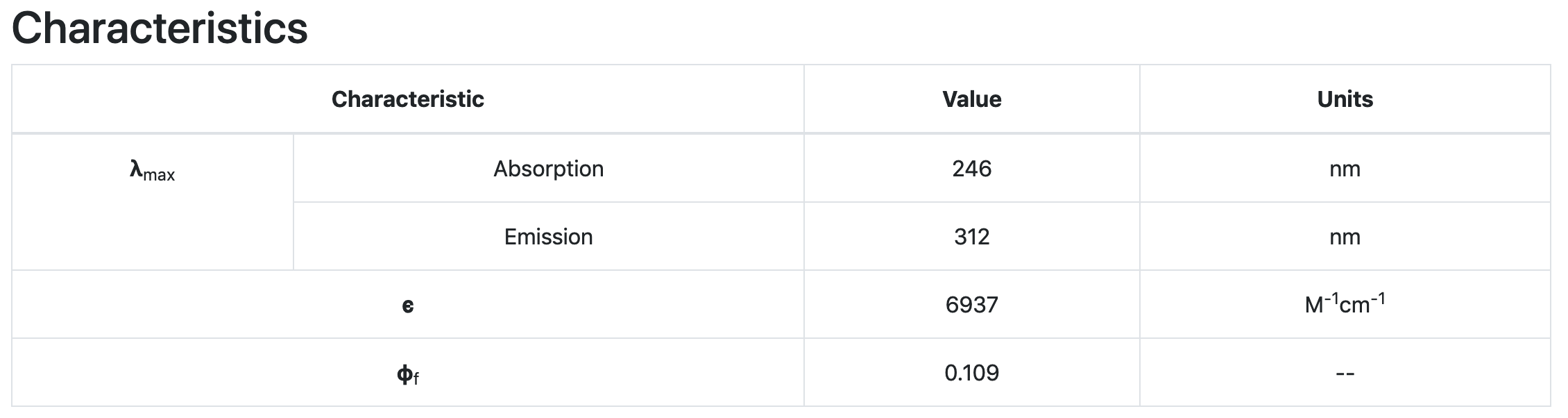
Below shows the spectra and characteristics prediction of an example molecule, inputted via our GUI.
- Python version 3.6.7 or later
- conda version 4.6.8 or later
- GitHub
You can execute the following commands from your computer's terminal application:
-
Either clone the SPEEDCOM repository:
git clone https://github.com/emissible/SPEEDCOM.gitor download the zip file:
curl -O https://github.com/emissible/SPEEDCOM/archive/master.zip -
cd SPEEDCOM -
conda env create -n speedcom_environment.yml -
conda activate speedcom -
cd speedcom && python runhtml.py -
Find the isomeric SMILES string for the moelcule you want the spectra for and input it into the GUI!
SPEEDCOM (master)
|---data
|---
|---doc
|---
|---speedcom
|---frontend
|---output
|--jquery.min.js
|--molecule_ex.png
|--spectra_ex.png
|--welcome.png
|---notebook_scripts
|--R2_plot.ipynb
|--encode_smiles.ipynb
|--rdkit_descriptor.ipynb
|--rdkit_exploration.ipynb
|--smiles_clean.ipynb
|--smiles_cnn.ipynb
|--to_help_write_tests.ipynb
|---saved_models
|--ems_dropna.best1.hdf5
|--epsilon.best_-1.hdf5
|--model_lstm_qy.json
|--model_smiles_cnn.json
|--model_smiles_ems_dropna.json
|--model_smiles_epsilon_lstm.json
|--smiles_wordmap.json
|--weights.best.hdf5
|--weight.qy_best.hdf5
|---tests
|---DATA_CLEAN_TEST_DIR
|--__init__.py
|--context,py
|--tests_NNModels.py
|--test_Prediction.py
|--test_dataUtils.py
|--test_data_extract.py
|--test_model_utils.py
|--test_readData_temp.tsv
|--test_speedcom.py
|--test_utilities.py
|--NNModels.py
|--Prediction.py
|--__init__.py
|--core.py
|--dataUtils.py
|--data_extract.py
|--matplotlibrc
|--model_utils.py
|--runhtml.py
|--speedcom.html
|--utilities.py
|--version.py
|--.coveragerc
|--.gitignore
|--.travis.yml
|--LICENSE
|--README.md
|--download.sh
|--numpyversion.py
|--requirements.txt
|--runtests.sh
|--setup.sh
|--speedcom_environment.yml
Any contributions to the project are warmly welcomed! If you discover any bugs, please report them in the issues section of this repository and we'll work to sort them out as soon as possible. If you have data that you think will be good to train our model on, please contact one of the authors.
Garrett B. Goh et al. 2018. SMILES2vec. In Proceedings of ACM SIGKDD Conference, London, UK, Aug, 2018 (KDD 2018), 8 pages
SPEEDCOM is licensed under the MIT license.