-
Running environment:
- The workflow was constructed based on the Linux system running the R v4.1.1.
-
Required software and versions:
install.packages(c("data.table", "glmnet", "MASS", "rrBLUP", "parallel",
"doParallel", "CompQuadForm", "circlize", "dplyr", "RcolorBrewer"))
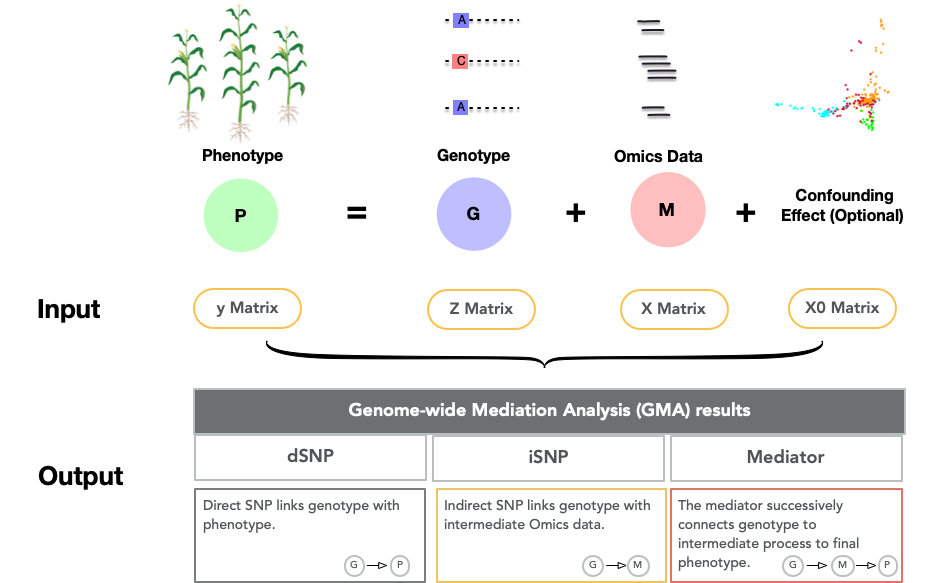
- y matrix (
input/y_matrix.txt):- A
n x 1matrix of phenotype values;nis the number of individuals.
- A
library("data.table")
y <- fread("input/y_matrix.txt", header=T,data.table=FALSE)
y = as.matrix(y)
dim(y) #271 1
head(y)
- Z matrix (
input/Z_matrix.txt):- The
n x mgenotype matrix of data;mis the number of bi-allelic SNP markers, coded as-1, 0, 1.
- The
Z <- fread("input/Z_matrix.txt", header=T,data.table=FALSE)
Z = as.matrix(Z)
dim(Z) #271 5000
Z[1:10, 1:10]
- X matrix (
input/X_matrix.txt):- The
n x kintermediate Omics matrix;kis the number of Omics traits. In the example, gene expression data (i.e., RNA-seq read counts ofkgenes) was used as the intermediate traits .
- The
X <- fread("input/X_matrix.txt", header=T,data.table=FALSE)
X = as.matrix(X)
dim(X) #271 1200
X[1:10, 1:10]
- X0 matrix (
input/X0_matrix.txt):- A matrix of confounding effects. In the example, three principal components calculated from the Z matrix were used to control population structure.
X0 <- fread("input/X0_matrix.txt", header=T,data.table=FALSE)
X0 = as.matrix(X0)
dim(X0) #271 3
head(X0)
# X0 = prcomp(Z)$x[,1:3] # use this line of code to calculate principal components if no X0_matrix.txt file provided
Several principal components can be used as the fixed effects to control population structure as the confounding effects if using a structured population.
source('lib/utils.R')
Z <- fread("input/Z_matrix.txt", header=T, data.table=FALSE)
Z = as.matrix(Z)
X0 <- getpca(Z, p=3) # here the first p=3 PCs were extracted.
#library(GMA)
library(glmnet)
library(MASS)
library(rrBLUP)
library(parallel)
library(doParallel)
library(CompQuadForm)
source('lib/highmed2019.r')
source('lib/fromSKAT.R')
source('lib/MedWrapper.R')
source('lib/reporters.R')
subX = X[, 1:100]
# run the fixed effect model that assign equal penalty on the two data types.
run_GMA(y, X0, subX, Z, ncores=10, model="MedFix_eq", output_folder="output/")
# run the fixed effect model that minimizes BIC
run_GMA(y, X0, subX, Z, ncores=10, model="MedFix_fixed", output_folder="output/")
# run the random effect model using linear kernel, and extract the model that minimizes BIC
run_GMA(y, X0, subX, Z, ncores=10, model="MedMix_linear", output_folder="output/")
# run the random effect model using shrink_EJ kernel, and extract the model that minimizes BIC
run_GMA(y, X0, subX, Z, ncores=10, model="MedMix_shrink", output_folder="output/")
library(circlize)
library(dplyr)
library("RColorBrewer")
gwas <- qGWAS(y, Z, plot=FALSE)
fwrite(gwas, "output/gwas_results.csv", sep=",", row.names = FALSE, quote=FALSE)
source("lib/circosplot.R")
circos_med(gwas_res="output/gwas_results.csv",
med_res="output/mediators_fixed_bic_trait_V1.csv",
dsnp_res="output/dsnps_fixed_bic_trait_V1.csv",
isnp_res="output/isnps_fixed_bic_trait_V1.csv",
chrlen="input/Chromosome_v4.txt",
gene_position= "input/gene_pos.csv",
out_tiff = "graphs/circos.tiff")
The outputs of the example data:
Direct SNPs identified using MedFix_eq and MedFix_fixed methods for trait V1. Note that only MedFix methods will report direct SNP.
output/dSNP_MedFix_eq_trait_V1.csvoutput/dSNP_MedFix_fixed_trait_V1.csv
The dSNP output files contain the following columns:
- snp: direct SNP;
- pval: p-value of effect from exposure to outcome;
- coef: SNP effect from exposure to outcome.
Indirect SNPs identified. Again, only MedFix methods will report direct SNPs.
output/iSNP_MedFix_eq_trait_V1.csvoutput/iSNP_MedFix_eq_trait_V1.csv
The iSNP output files contain the following columns:
- medi: mediator gene under control by the iSNP;
- snps_for_medi: indirect SNPs for the corresponding mediator;
- coef: effect from exposure to mediator.
The non-adjusted mediators detected by different methods of MedFix_BIC, MedFix_0.5, MedMixed_Linear, and MedMixed_Shrink for trait V1.
output/mediator_MedFix_eq_trait_V1.csvoutput/mediator_MedFix_eq_trait_V1.csvoutput/mediator_MedMix_linear_trait_V1.csvoutput/mediator_MedFix_eq_trait_V1.csv:
The mediator output files contain the following columns:
- id: mediator gene id;
- e2m: p-value of effect from exposure to mediator;
- m2y: p-value of effect from mediator to outcome;
- e2m2y: maximum value between e2m and m2y;
- padj: adjusted p-value;
- coef: product of effect from exposure to mediator and effect from mediator to outcome.
- In the circos plots, the outermost circular track represents the ten chromosomes;
- The next inner track shows the GWAS results, with two circular blue dashed lines indicating -log(p-value) of 5 and 10 and the red lines denoting the position of direct SNPs;
- The next inner track shows the relative positions of identified mediator genes with different genes represented by different colors; the lines in the innermost circle connects mediators with their corresponding indirect SNPs.
- Zhang, Qi. "High-dimensional mediation analysis with applications to causal gene identification." Statistics in Biosciences (2021): 1-20.
- Yang, Zhikai, Gen Xu, Qi Zhang, Toshihiro Obata, and Jinliang Yang. "Genome-wide mediation analysis: an empirical study to connect phenotype with genotype via intermediate transcriptomic data in maize." Genetics 221, no. 2 (2022): iyac057.
It is a free and open source software, licensed under GPLv3.