This project aims to provide a simple way to select models (or specifically classifiers).
- You can organize classifiers, preprocessors and parameter spaces.
- You can easily ensemble them according to cases.
- Lastly, you can create a experiment manager and run in one call.
- Add path of
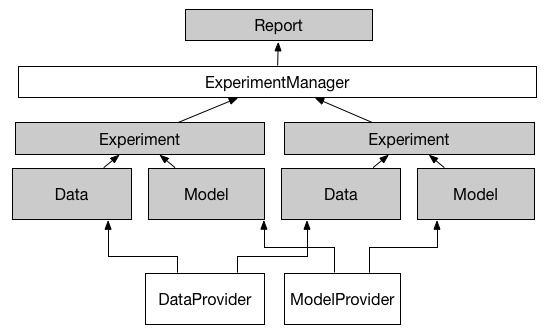
MatExManager. - Prepare basic elements of your models (classifiers, preprocessors and parameter spaces) in
ClassifierProvider,PreprocessorProviderandModelParamProvider. - Create your
ModelProviderwhere you can ensemble classifiers, preprocessors and parameter spaces. Alternatively, there is a minimal demo to do this. - Provide your data inside
- Run all methods provided in
ModelProvideror see more in demo:
EM = ExperimentManager ( datasetName, options ); % Init with data set and options.
EM.setup(); % set up
EM.runAll(); % Run all required methods.
results = EM.outputResults(); % Get formatted results.This class implement two methods inside: getNames and getModelByName. Practically, you need to create a subclass of ModelProvider and implement all abstract methods. You can modify the file, ModelProvider.m, to add your own models.
We want to provide a model who uses SVM as classifier, process data into RBF-kernel matrix.
Easily, you can see the simple demo in DemoModelProvider where all functions you need to modify is enclosed here. However, we recommend a more organized way to store these functions.
Step 1: define the string name of the model as svm_rbf.
function names = getNames ()
names = {'svm_rbf', % SVM classifier with RBF kernel
};
endStep 2: provide the elements of the model.
function [preprocessor, classifier, modelParam] = getModelByName ( name, options )
switch name
case 'svm_rbf'
modelParam = ModelParamProvider.svm_rbf(options);
classifier = @ClassifierProvider.svm;
preprocessor = @PreprocessorProvider.kernel_preprocessor;
end
endThe three outputs should be formatted as
preprocessor(function handler):newdata = fun (data)where structnewdatashould contain three fields:X,Y,test_X,test_Yas training data&label, testing datta&label.classifier(function handler):[ W, test_err, train_err ] = fun (data)wheredatais a struct with fields like:data.X.K, data.Y, data.test_X.K, data.test_Y, data.options;Wis model, e.g. matrix of classifier coeficients.test_err,train_err: test/train error rate on the test set.
modelParam(ModelParamobject): SeeModelProviderfor how to generate a model parameter space easily.
A class to provide data. You need to create a subclass of DataProvider and implement all abstract methods. See data provider demo.
function loaded = load_from_file (DP)
% Load from file and return data in struct 'loaded'.
% You can customize the function to adapt your file format.
...
end
function [X, test_X, Y, test_Y] = process_data (DP, ds)
% You can customize the function to adapt your data format.
% For example, you can slice the data dimension.
...
endThis class provides sets of preprocessors for feature extraction, kernel computing and etc. An example computing kernel matrixes:
function newdata = kernel_preprocessor (data)
% cell array of data vectors -> kernel matrix.
% Kernel function handler
ker_fh = @(x1, x2) exp(-gam* sum((x1 - x2).^2));
% compute kernel
[ newdata.X, newdata.test_X, newdata.Y, newdata.test_Y ] = ...
compute_kernel ( ker_fh, data );
endIt is noticable that you have to make the data newdata.X as a cell array or a struct containing a kernel matrix. This is because the experiment manager can only make cross-validation partition available for these two formats.
To make different classifier adapt to the experiment manager, you need to write a function to make the transformation. An example of SVM:
function [ W, test_err, train_err ] = svm (data)
% process options
if isfield(data, 'options'); options = data.options; else; options = []; end;
[C] = process_options (options, 'C', 1);
% Call the real SVM
[ test_err, train_err, W ] = svm_none ( data.X.K, data.Y, data.test_X.K, data.test_Y, ...
struct('C', C) );
endThis class provide static methods to return ModelParam objects which enclose the whole parameter space for model selection. A simple demo:
function [ modelParam ] = svm_rbf ( options )
% Create a ModelParam with parameter space. Format: {'name', range, 'name', range, ...}
modelParam = ModelParam({'C', power(10, -4:5), ...
'gam', power(10, 0:-1:-4)});
endwhere we yield two parameter spaces named C and gam.