This code has been developed by Ali Davariashtiyani. (LinkedIn, Scholar)
This code has been developed under the support of the National Science Foundation (NSF) award number DMR-2119308.
Please cite the following paper if you use any part of our code or data:
Davariashtiyani, A., Kadkhodaie, Z. & Kadkhodaei, S. Predicting synthesizability of crystalline materials via deep learning. Commun Mater 2, 115 (2021). https://doi.org/10.1038/s43246-021-00219-x
The data files for this paper is available at https://uofi.app.box.com/s/7o39i8jt4ve7s5loo0twlub06k3d7np3
Ali Davariashtiyani, Sara Kadkhodaei. Formation energy prediction of crystalline compounds using deep convolutional network learning on voxel image representation. Commun Mater 2, 105 (2023). https://doi.org/10.1038/s43246-023-00433-9
The data files for this paper is available at https://uofi.box.com/s/ka69pgrpgn9gkauz912urcurj31xmlfm
Tested on Ubuntu 20.04
Navigating to the desired directory in the terminal:
cd [/navigate/to/directory/]Cloning the repository and entering the directory:
git clone https://github.com/kadkhodaei-research-group/XIE-SPP.git
cd XIE-SPP/We highly encourage you to use a conda envirenment. If you don't have conda installed on your machine, you can skip this step:
conda create -n synthesizability python=3.7.3 -y
conda activate synthesizabilityInstalling TensorFlow on Ubuntu:
***Installing the GPU version:
conda install -c anaconda keras-gpu -y
***Installing the CPU version:
conda install -c anaconda keras -yInstalling TensorFlow on Mac M1:
miniforge3 is required.
conda install -c apple tensorflow-deps -y
pip install tensorflow-macos
pip install tensorflow-metalInstalling the Crystal Image Encoder for Synthesis & Property Prediction (XIE-SPP) package and it's dependencies:
python setup.py installLoading the model:
from xiespp.synthesizability_1 import synthesizability
samples = synthesizability.get_test_samples('GaN')
samplesOutput:
['XIE-SPP/finalized_results/explore_structures/cif/GaN/GaN_9.cif',
'XIE-SPP/finalized_results/explore_structures/cif/GaN/GaN_12.cif']
Evaluation: (Input can be CIF files or ASE Atoms Objects)
synthesizability.synthesizability_predictor(samples)The output is the synthesizability likelihood of the input list:
2/2 [==============================] - 2s 783ms/step
array([0.00200533, 0.9643494 ], dtype=float32)
We highly encourage the use of this version of the model. This version is more robust, accurate and faster than the previous version.
from xiespp import synthesizability, get_test_samples
# or
# from xiespp import synthesizability_2 as synthesizability
samples = get_test_samples('CSi')
# The input to the model can in formats like: CIF, POSCAR, PyMatGen Structure, ASE Atoms
model = synthesizability.SynthesizabilityPredictor()
model.predict(samples)from xiespp import formation_energy, get_test_samples
model = formation_energy.FormationEnergyPredictor()
# As an example we take the samples from MP database
from mp_api.client import MPRester
api_key = '__YOUR_API_KEY__'
mpr = MPRester(api_key)
search = mpr.summary.search(chemsys="Si-O")
data_input = search[:10]
# As practice instead of directly passing the data to the model we first generate the Image Generator Object
gen = formation_energy.prepare_data(
data_input=data_input,
# input_format= 'vasp', # Use this if the format of the file is not CIF file
)
model = formation_energy.FormationEnergyPredictor()
yp = model.predict(gen, return_all_ensembles=True)Data files available at https://uofi.app.box.com/s/7o39i8jt4ve7s5loo0twlub06k3d7np3
Untarring the files to the data folder:
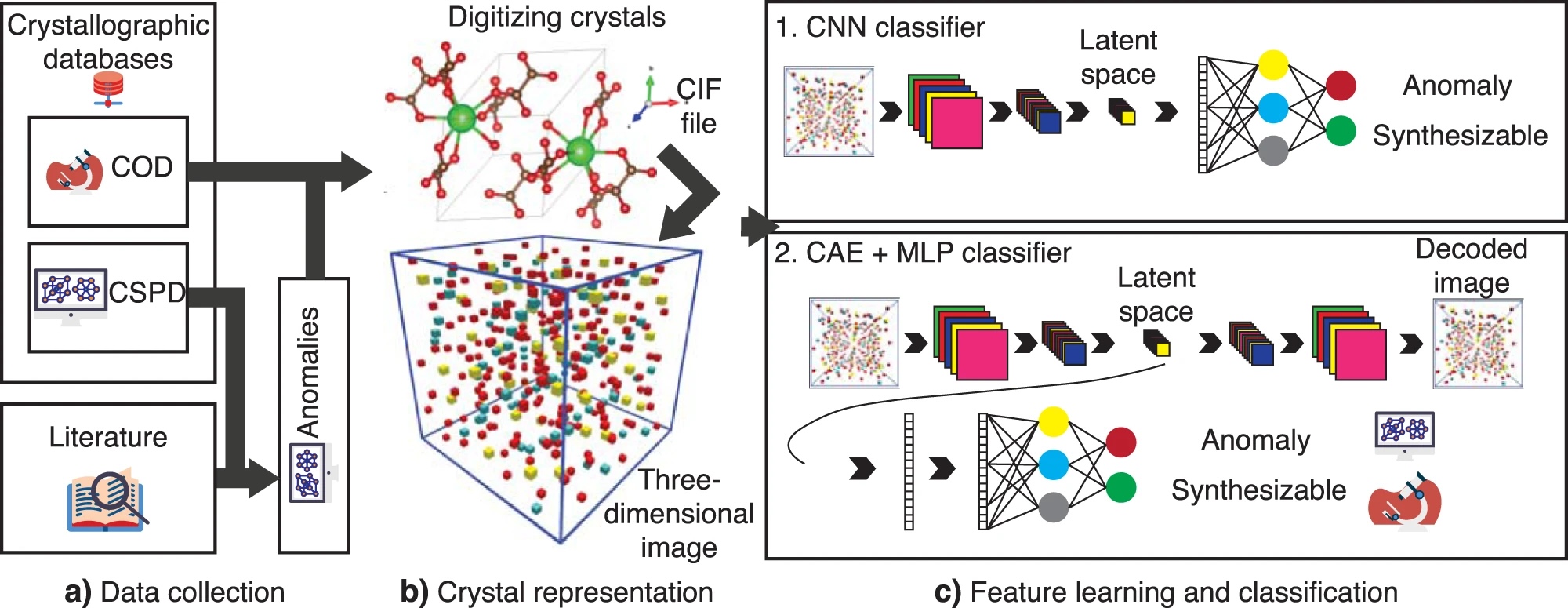
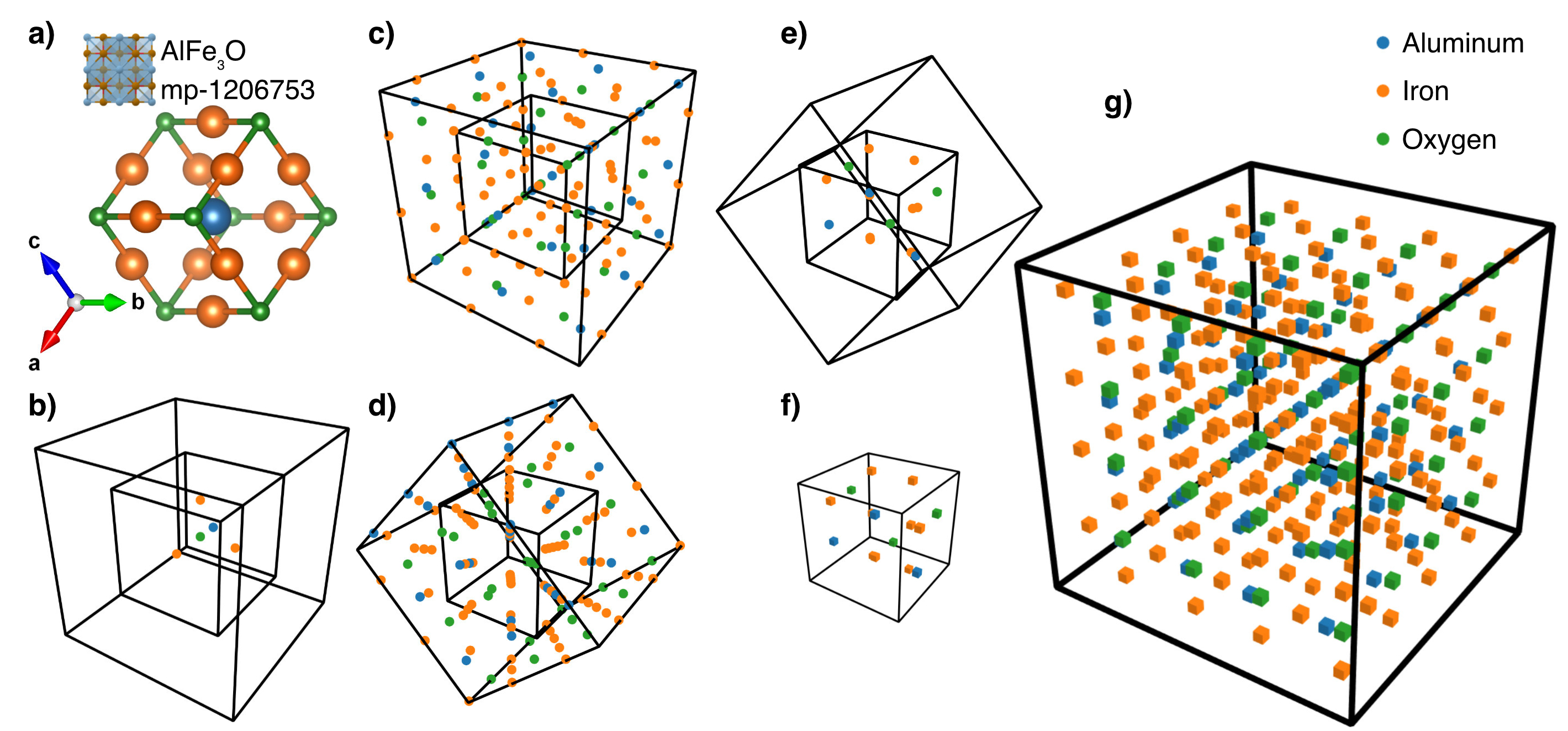
tar -xvzf Data.joined.tar.gzAll the CIF files are collected from the Materials Project, COD and CSPD databases. The selected data fed to the model is available in the unwrapped data folder.
All the used data is already prepared in the unwrapped folder. However, for reproducing everything again, follow the steps: Set the data folders in the config.py file.
- Prepare the positive set: Obtain the COD:
mkdir data/data_bases/
wget http://www.crystallography.net/archives/cod-cifs-mysql.tgz
tar -xvzf cod-cifs-mysql.tgzFollow the guideline to create the SQL file.
Run the entire positive_data_preparation.ipynb file.
- According to the prepared guideline to prepare the CSPD data set and run anomaly_generation.ipynb to prepare the negative set
- Create the image files by running data_set_selections.ipynb
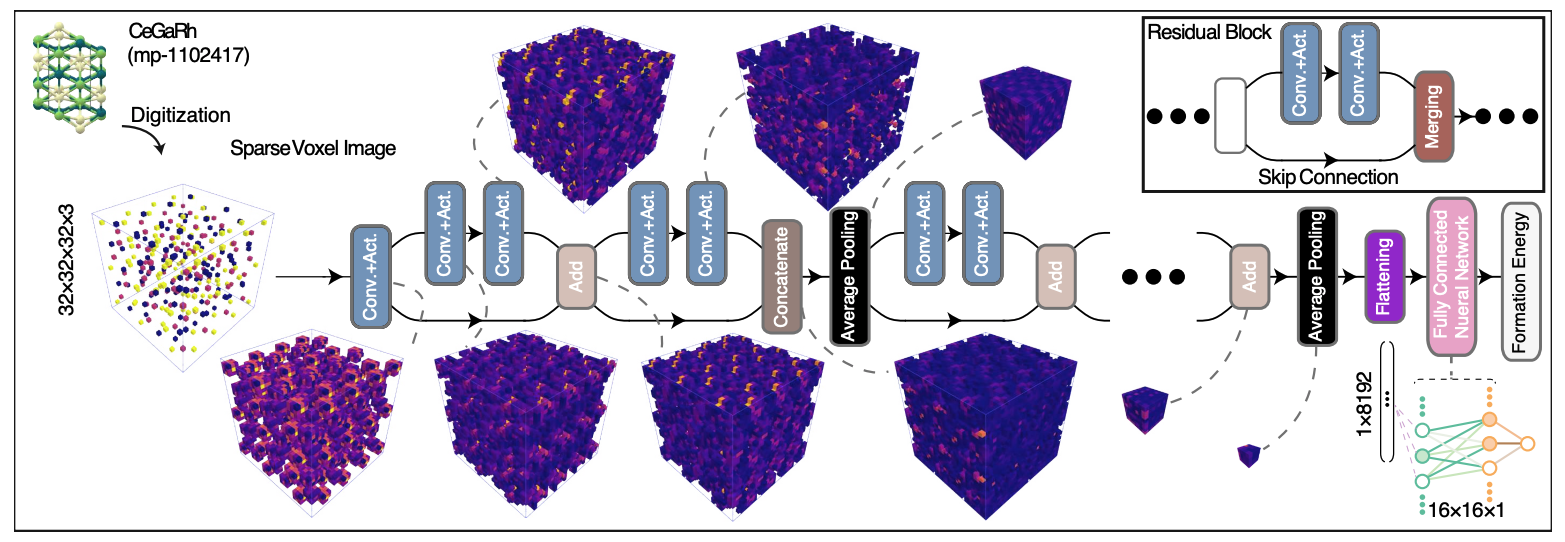
Re-training formation energy prediction: model7_training.ipynb
Image creation package: CVR