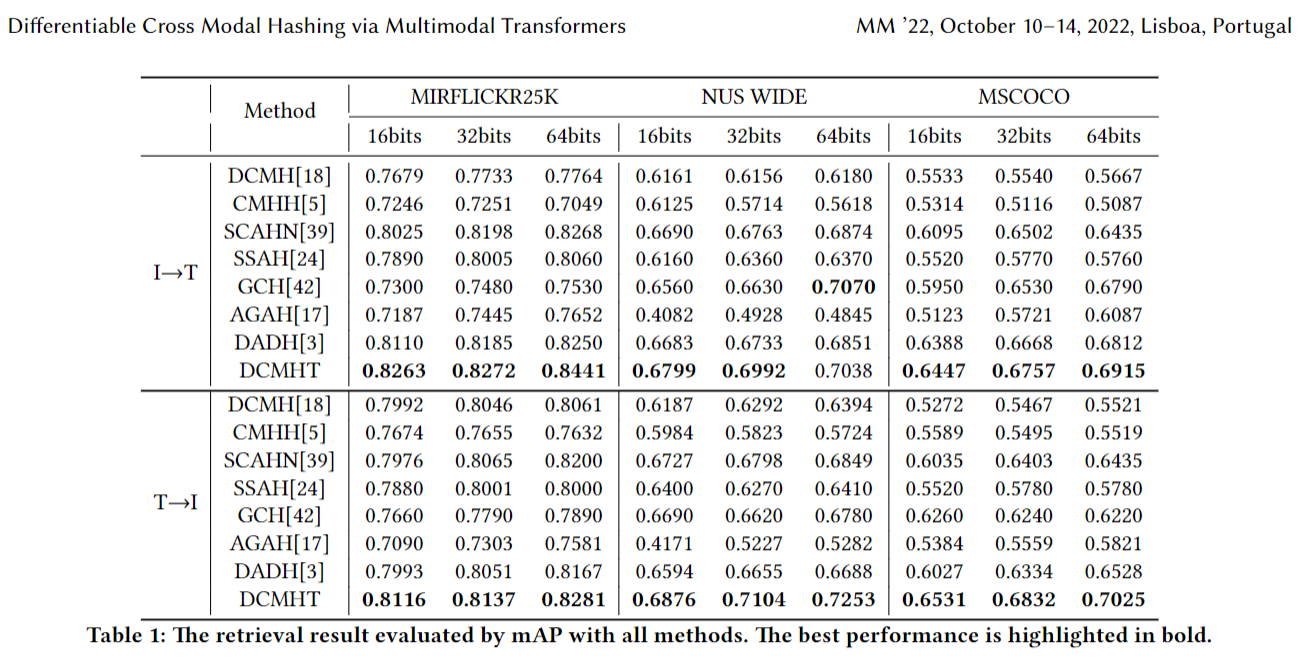
Differentiable Cross Modal Hashing via Multimodal Transformers paper
This project has been moved to clip-based-cross-modal-hash
The main architecture of our method.
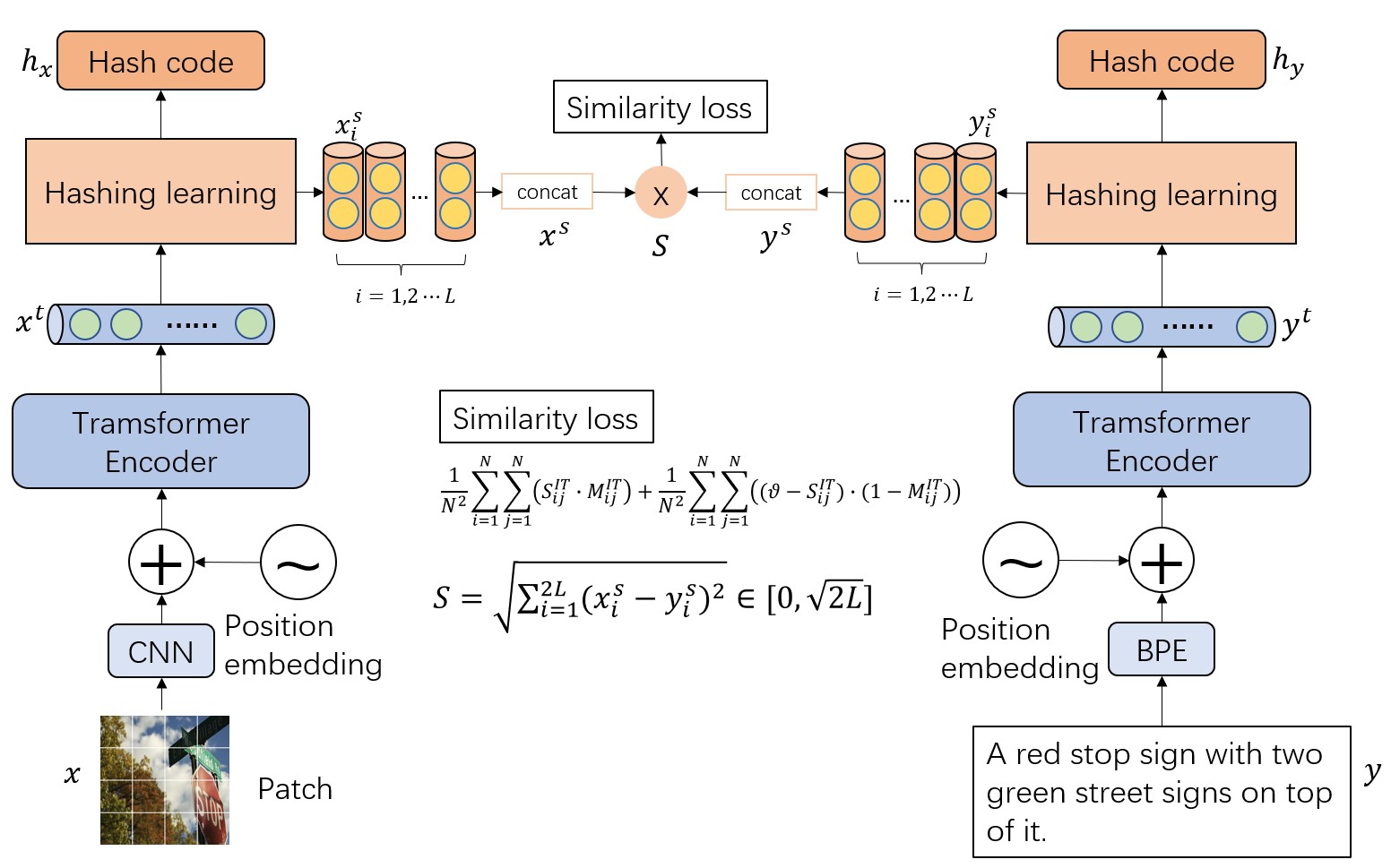
We propose a selecting mechanism to generate hash code that will transfor the discrete space into a continuous space. Hash code will be encoded as a seires of 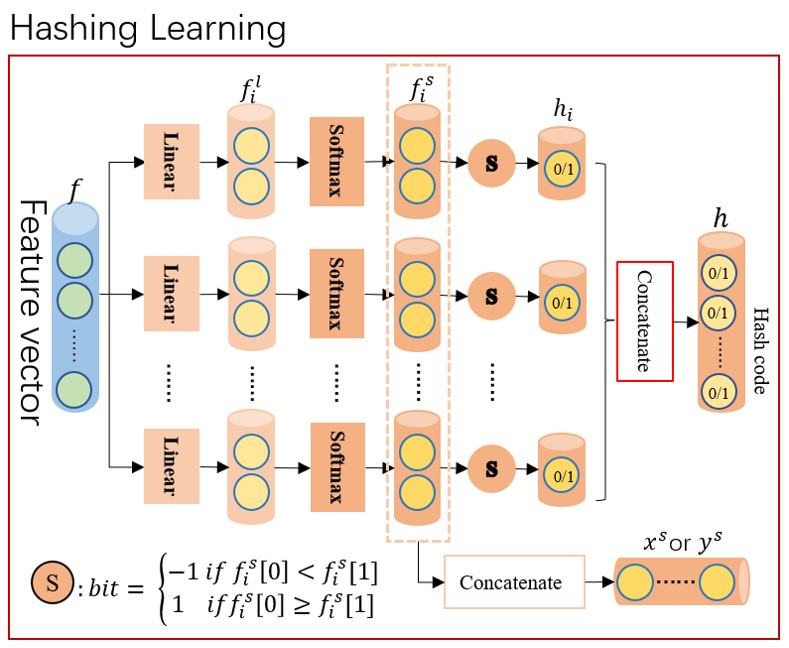
We use python to build our code, you need to install those package to run
- pytorch 1.9.1
- sklearn
- tqdm
- pillow
Before training, you need to download the oringal data from coco(include 2017 train,val and annotations), nuswide(include all), mirflickr25k(include mirflickr25k and mirflickr25k_annotations_v080), then use the "data/make_XXX.py" to generate .mat file
For example:
cd COCO_DIR # include train val images and annotations files
mkdir mat
cp DCMHT/data/make_coco.py mat
python make_coco.py --coco-dir ../ --save-dir ./
After all mat file generated, the dir of dataset will like this:
dataset
├── base.py
├── __init__.py
├── dataloader.py
├── coco
│ ├── caption.mat
│ ├── index.mat
│ └── label.mat
├── flickr25k
│ ├── caption.mat
│ ├── index.mat
│ └── label.mat
└── nuswide
├── caption.txt # Notice! It is a txt file!
├── index.mat
└── label.mat
Pretrained model will be found in the 30 lines of CLIP/clip/clip.py. This code is based on the "ViT-B/32".
You should copy ViT-B-32.pt to this dir.
After the dataset has been prepared, we could run the follow command to train.
python main.py --is-train --hash-layer select --dataset coco --caption-file caption.mat --index-file index.mat --label-file label.mat --similarity-function euclidean --loss-type l2 --vartheta 0.75 --lr 0.0001 --output-dim 64 --save-dir ./result/coco/64 --clip-path ./ViT-B-32.pt --batch-size 256
inproceedings{10.1145/3503161.3548187,
author = {Tu, Junfeng and Liu, Xueliang and Lin, Zongxiang and Hong, Richang and Wang, Meng},
title = {Differentiable Cross-Modal Hashing via Multimodal Transformers},
year = {2022},
booktitle = {Proceedings of the 30th ACM International Conference on Multimedia},
pages = {453–461},
numpages = {9},
}
2023/03/01
I find figure 1 with the wrong formula for the vartheta, the right one is the function (10). It has been published, so I can't fix it.