⚗️ Status: This project is still in alpha, and the API may change without warning.
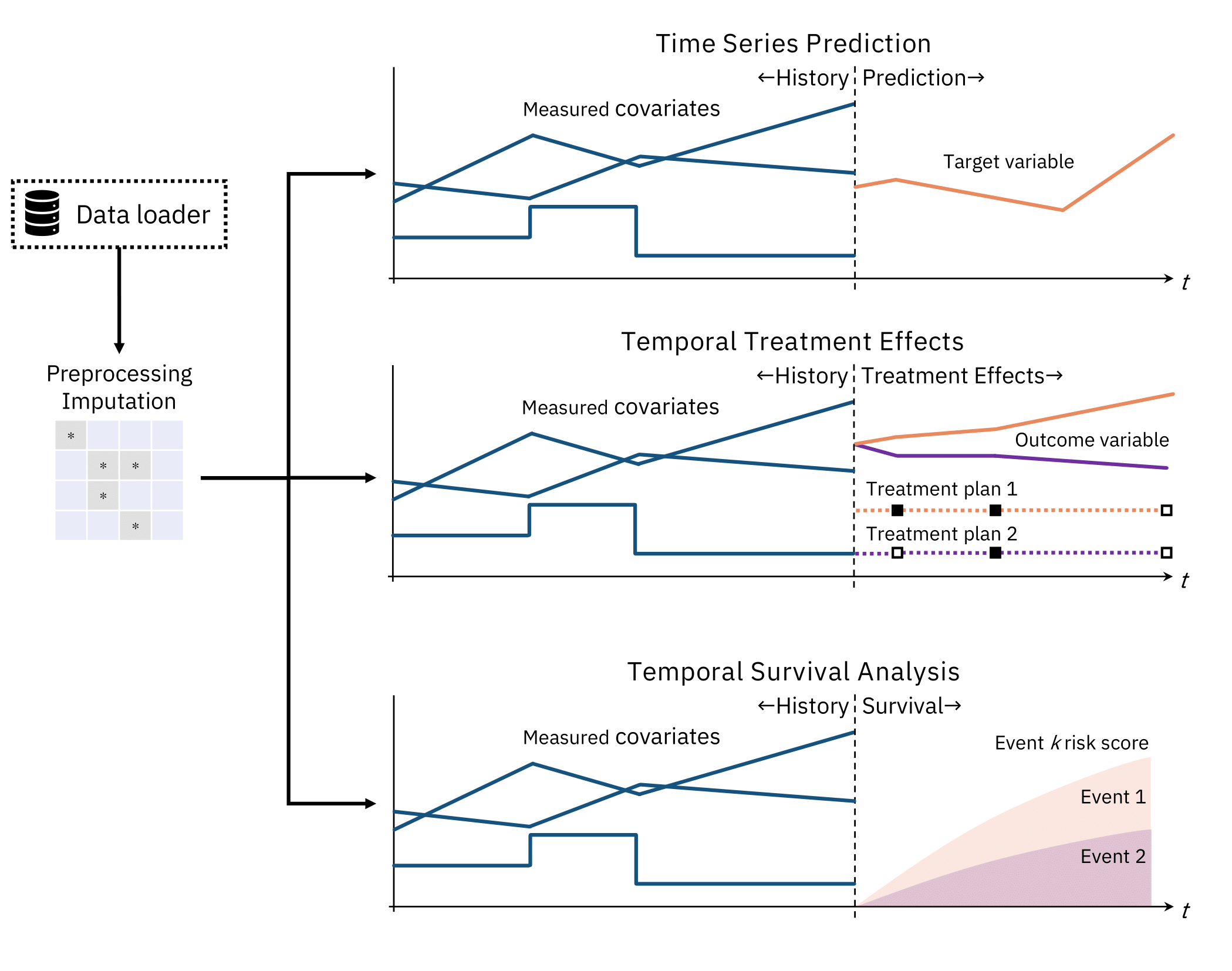
TemporAI is a Machine Learning-centric time-series library for medicine. The tasks that are currently of focus in TemporAI are: time-series prediction, time-to-event (a.k.a. survival) analysis with time-series data, and counterfactual inference (i.e. [individualized] treatment effects).
In future versions, the library also aims to provide the user with understanding of their data, model, and problem, through e.g. integration with interpretability methods.
Key concepts:
$ pip install temporaior from source, using
$ pip install .- List the available plugins
from tempor.plugins import plugin_loader
print(plugin_loader.list())- Use an imputer
from tempor.utils.dataloaders import SineDataLoader
from tempor.plugins import plugin_loader
dataset = SineDataLoader(with_missing=True).load()
static_data_n_missing = dataset.static.dataframe().isna().sum().sum()
temporal_data_n_missing = dataset.time_series.dataframe().isna().sum().sum()
print(static_data_n_missing, temporal_data_n_missing)
assert static_data_n_missing > 0
assert temporal_data_n_missing > 0
# Load the model:
model = plugin_loader.get("preprocessing.imputation.temporal.bfill")
# Train:
model.fit(dataset)
# Impute:
imputed = model.transform(dataset)
static_data_n_missing = imputed.static.dataframe().isna().sum().sum()
temporal_data_n_missing = imputed.time_series.dataframe().isna().sum().sum()
print(static_data_n_missing, temporal_data_n_missing)
assert static_data_n_missing == 0
assert temporal_data_n_missing == 0- Use a classifier
from tempor.utils.dataloaders import SineDataLoader
from tempor.plugins import plugin_loader
dataset = SineDataLoader().load()
# Load the model:
model = plugin_loader.get("prediction.one_off.classification.nn_classifier", n_iter=50)
# Train:
model.fit(dataset)
# Predict:
prediction = model.predict(dataset)- Use a regressor
from tempor.utils.dataloaders import SineDataLoader
from tempor.plugins import plugin_loader
dataset = SineDataLoader().load()
# Load the model:
model = plugin_loader.get("prediction.one_off.regression.nn_regressor", n_iter=50)
# Train:
model.fit(dataset)
# Predict:
prediction = model.predict(dataset)- Benchmark models Classification task
from tempor.benchmarks import benchmark_models
from tempor.plugins import plugin_loader
from tempor.plugins.pipeline import Pipeline
from tempor.utils.dataloaders import SineDataLoader
testcases = [
(
"pipeline1",
Pipeline(
[
"preprocessing.scaling.static.static_minmax_scaler",
"prediction.one_off.classification.nn_classifier",
]
)({"nn_classifier": {"n_iter": 10}}),
),
(
"plugin1",
plugin_loader.get("prediction.one_off.classification.nn_classifier", n_iter=10),
),
]
dataset = SineDataLoader().load()
aggr_score, per_test_score = benchmark_models(
task_type="classification",
tests=testcases,
data=dataset,
n_splits=2,
random_state=0,
)
print(aggr_score)- Serialization
from tempor.utils.serialization import load, save
from tempor.plugins import plugin_loader
# Load the model:
model = plugin_loader.get("prediction.one_off.classification.nn_classifier", n_iter=50)
buff = save(model) # Save model to bytes.
reloaded = load(buff) # Reload model.
# `save_to_file`, `load_from_file` also available in the serialization module.Prediction where targets are static.
- Classification (category:
prediction.one_off.classification)
| Name | Description | Reference |
|---|---|---|
nn_classifier |
Neural-net based classifier. Supports multiple recurrent models, like RNN, LSTM, Transformer etc. | --- |
ode_classifier |
Classifier based on ordinary differential equation (ODE) solvers. | --- |
cde_classifier |
Classifier based Neural Controlled Differential Equations for Irregular Time Series. | Paper |
laplace_ode_classifier |
Classifier based Inverse Laplace Transform (ILT) algorithms implemented in PyTorch. | Paper |
- Regression (category:
prediction.one_off.regression)
| Name | Description | Reference |
|---|---|---|
nn_regressor |
Neural-net based regressor. Supports multiple recurrent models, like RNN, LSTM, Transformer etc. | --- |
ode_regressor |
Regressor based on ordinary differential equation (ODE) solvers. | --- |
cde_regressor |
Regressor based Neural Controlled Differential Equations for Irregular Time Series. | Paper |
laplace_ode_regressor |
Regressor based Inverse Laplace Transform (ILT) algorithms implemented in PyTorch. | Paper |
Prediction where targets are temporal (time series).
- Classification (category:
prediction.temporal.classification)
| Name | Description | Reference |
|---|---|---|
seq2seq_classifier |
Seq2Seq prediction, classification | --- |
- Regression (category:
prediction.temporal.regression)
| Name | Description | Reference |
|---|---|---|
seq2seq_regressor |
Seq2Seq prediction, regression | --- |
Risk estimation given event data (category: time_to_event)
| Name | Description | Reference |
|---|---|---|
dynamic_deephit |
Dynamic-DeepHit incorporates the available longitudinal data comprising various repeated measurements (rather than only the last available measurements) in order to issue dynamically updated survival predictions | Paper |
ts_coxph |
Create embeddings from the time series and use a CoxPH model for predicting the survival function | --- |
ts_xgb |
Create embeddings from the time series and use a SurvivalXGBoost model for predicting the survival function | --- |
Treatment effects estimation where treatments are a one-off event.
- Regression on the outcomes (category:
treatments.one_off.regression)
| Name | Description | Reference |
|---|---|---|
synctwin_regressor |
SyncTwin is a treatment effect estimation method tailored for observational studies with longitudinal data, applied to the LIP setting: Longitudinal, Irregular and Point treatment. | Paper |
Treatment effects estimation where treatments are temporal (time series).
- Classification on the outcomes (category:
treatments.temporal.classification)
| Name | Description | Reference |
|---|---|---|
crn_classifier |
The Counterfactual Recurrent Network (CRN), a sequence-to-sequence model that leverages the available patient observational data to estimate treatment effects over time. | Paper |
- Regression on the outcomes (category:
treatments.temporal.regression)
| Name | Description | Reference |
|---|---|---|
crn_regressor |
The Counterfactual Recurrent Network (CRN), a sequence-to-sequence model that leverages the available patient observational data to estimate treatment effects over time. | Paper |
- Static data (category:
preprocessing.imputation.static)
| Name | Description | Reference |
|---|---|---|
static_imputation |
Use HyperImpute to impute both the static and temporal data | Paper |
- Temporal data (category:
preprocessing.imputation.temporal)
| Name | Description | Reference |
|---|---|---|
ffill |
Propagate last valid observation forward to next valid | --- |
bfill |
Use next valid observation to fill gap | --- |
- Static data (category:
preprocessing.scaling.static)
| Name | Description | Reference |
|---|---|---|
static_standard_scaler |
Scale the static features using a StandardScaler | --- |
static_minmax_scaler |
Scale the static features using a MinMaxScaler | --- |
- Temporal data (category:
preprocessing.scaling.temporal)
| Name | Description | Reference |
|---|---|---|
ts_standard_scaler |
Scale the temporal features using a StandardScaler | --- |
ts_minmax_scaler |
Scale the temporal features using a MinMaxScaler | --- |
- Data Format
- Datasets
- Data Loaders
- Plugins
- Imputation
- Scaling
- Prediction
- Time-to-event Analysis
- Treatment Effects
- Pipeline
- Plugins
See the project documentation here.
Install the testing dependencies using
pip install .[dev]The tests can be executed using
pytest -vsxIf you use this code, please cite the associated paper:
@article{saveliev2023temporai,
title={TemporAI: Facilitating Machine Learning Innovation in Time Domain Tasks for Medicine},
author={Saveliev, Evgeny S and van der Schaar, Mihaela},
journal={arXiv preprint arXiv:2301.12260},
year={2023}
}