This Git repository contains codes for the 'On the influence of over-parameterization in manifold based surrogates and deep neural operators' paper which can be found here: https://www.sciencedirect.com/science/article/abs/pii/S0021999123001031 (JPC) or https://arxiv.org/abs/2203.05071 (arXiv)
Authors: Katiana Kontolati, Somdatta Goswami, Michael D. Shields, George Em Karniadakis
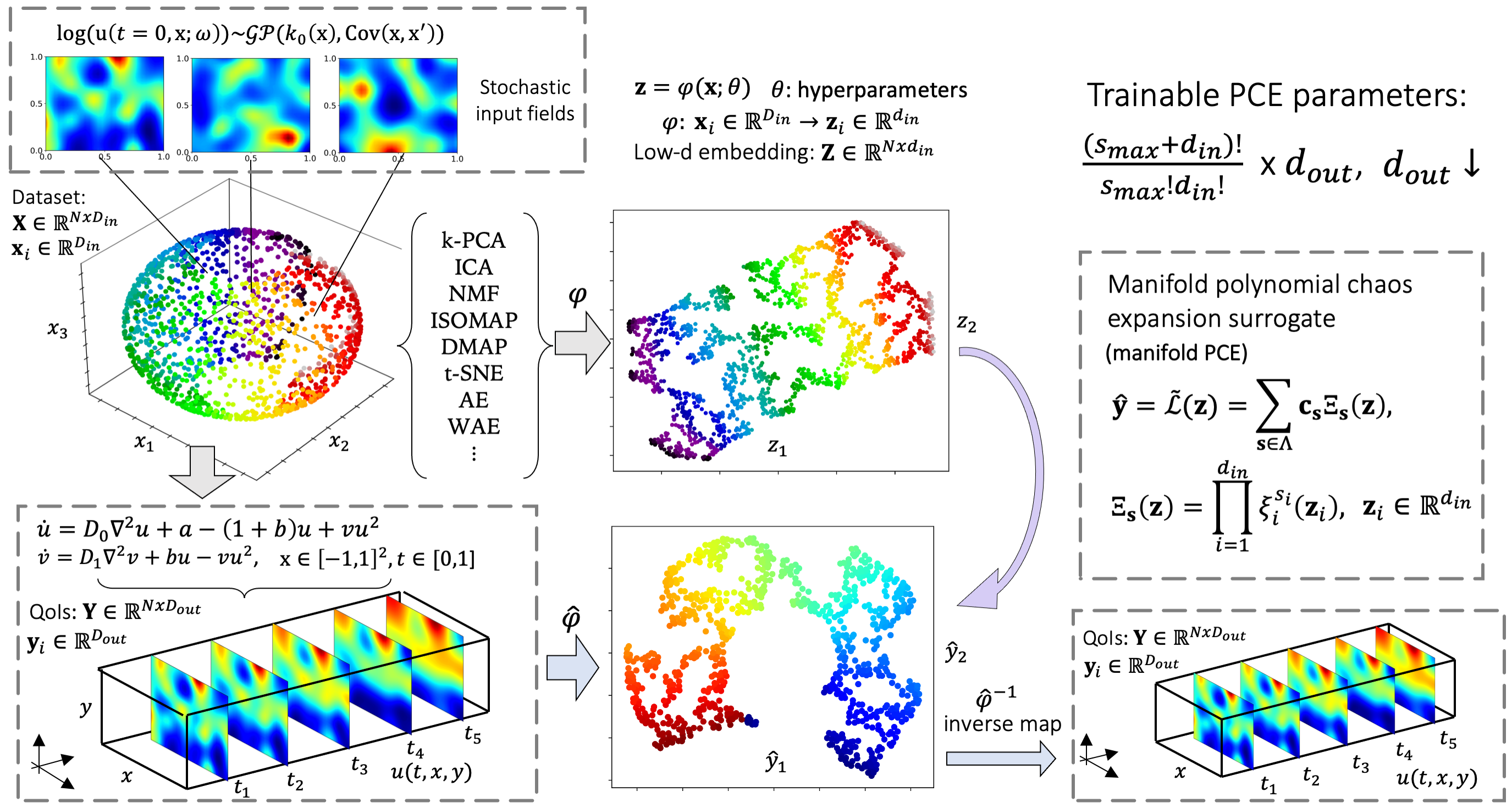
- Manifold PCE or mPCE approximates mappings via the identification of low-dimensional embeddings of input and output functions and the construction of a polynomial-based surrogate.
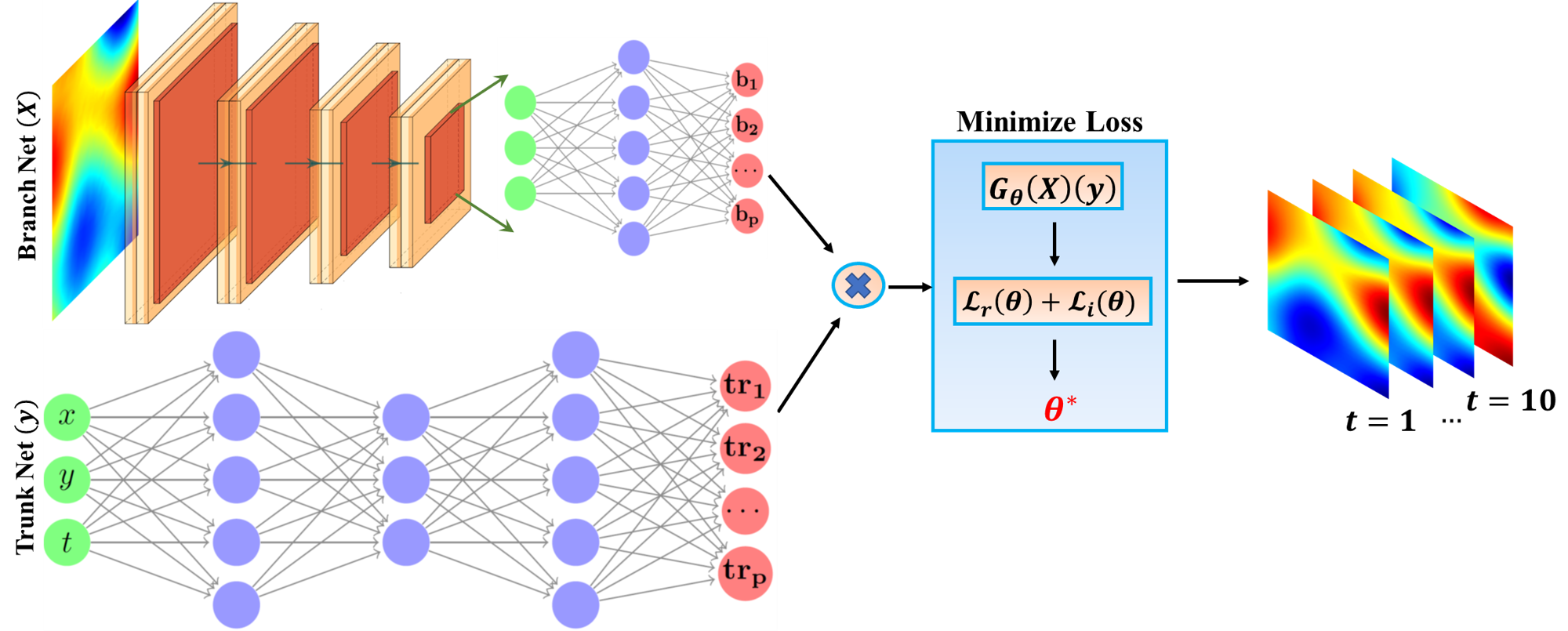
- DeepONet is a deep neural operator model which allows the construction of mapping between infinite dimensional functions via the use of deep neural networks (DNNs).
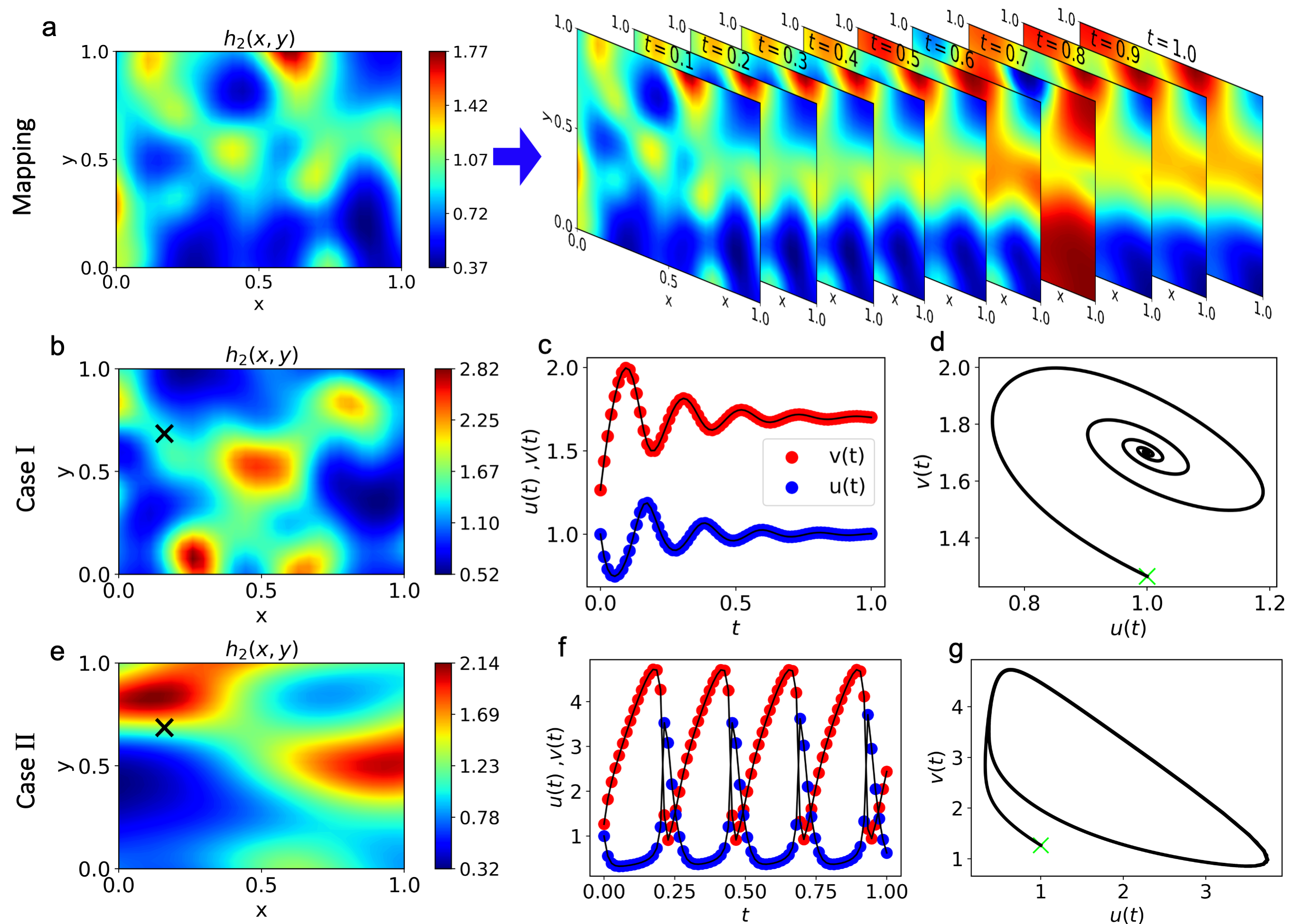
- The Brusselator diffusion-reaction dynamical system is studied, which describes an autocatalytic chemical reaction between two substances.
- The objective is to approximate the mapping between high-dimensional stochastic initial fields and the evolution of the system across time and space (first row below). The model response is learned for two dynamical states, when the system reaches a fixed point in the phase space (second row) and when it reaches a limit cycle (third row).
- We explore the capabilities of the studied models and test them for various regression tasks including their extrapolation/generalization ability (performance to out-of-distribution data), robustness to noise, ability to handle complex stochastic inputs and highly nonlinear mappings.
-
data- contains files with the input random field data used to generate the train and test data of the model -
utils- contains python scripts necessary for implementing the surrogate modeling tasks (loading data, plotting etc.) -
main_{}.py/main_{}.ipynb- contains python scripts and notebooks for implementing the proposed approaches
To generate the train/test data for the Brusselator model simply run the generate_data.py script. This will save the generated dataset inside folder data/. The py-pde package is used for solving the model.
To clone and use this repository, run the following terminal commands:
1. Create an Anaconda Python 3.7 virtual environment:
conda create -n surrogate-overparam python==3.7
conda activate surrogate-overparam
2. Clone the repo:
To clone and use this repository, run the following terminal commands:
git clone https://github.com/katiana22/surrogate-overparameterization.git
3. Install dependencies:
cd surrogate-overparameterization
pip install -r requirements.txt
If you find this GitHub repository useful for your work, please consider citing this work:
@article{kontolati2023influence,
title={On the influence of over-parameterization in manifold based surrogates and deep neural operators},
author={Kontolati, Katiana and Goswami, Somdatta and Shields, Michael D and Karniadakis, George Em},
journal={Journal of Computational Physics},
volume={479},
pages={112008},
year={2023},
publisher={Elsevier}
}
For more information or questions please contact us at: