a Customized Proteogenomic Workflow for Neoantigen Prediction and Selection
If you find it helpful, please consider citing our paper.
Li, Y., Wang, G., Tan, X. et al. ProGeo-neo: a customized proteogenomic workflow for neoantigen prediction and selection. BMC Med Genomics 13, 52 (2020). https://doi.org/10.1186/s12920-020-0683-4
ProGeo-neo requires a Linux operation system (centos6) with Python (V2.7) , Perl and Java installed.
In order to run normally, some third-party software such as BWA ,Gatk,and Annovar need extra databases. Here we provided these files in the reference_files, such as Hg38.fasta. In addition, during annotating genetic variants, annovar software needs lots of databases including: refGene, ensGene, cytoBand, avsnp147, dbnsfp30a, MT_ensGeneMrna, refGeneWithVerMrna, etc. of hg 38, putting them into humandb folder for the sake of convenience.
cd ProGeo-neo
bash start.sh
Users with root privileges can ignore the following:
chmod 755 soft/bwa/bwa
chmod 755 soft/samtools/samtools
chmod 755 soft/bcftools/bcftools
chmod 755 soft/gatk/gatk
chmod 755 soft/annovar/convert2annovar.pl
chmod 755 soft/annovar/table_annovar.pl
chmod 755 soft/annovar/annotate_variation.plpython get_variant-fasta.py /path/to/RNA-seq1_1.fastq /path/to/RNA-seq1_2.fastqeg: python get_variant-fasta.py test/rna/rnaseq-sample1_1.fastq test/rna/rnaseq-sample1_2.fastq
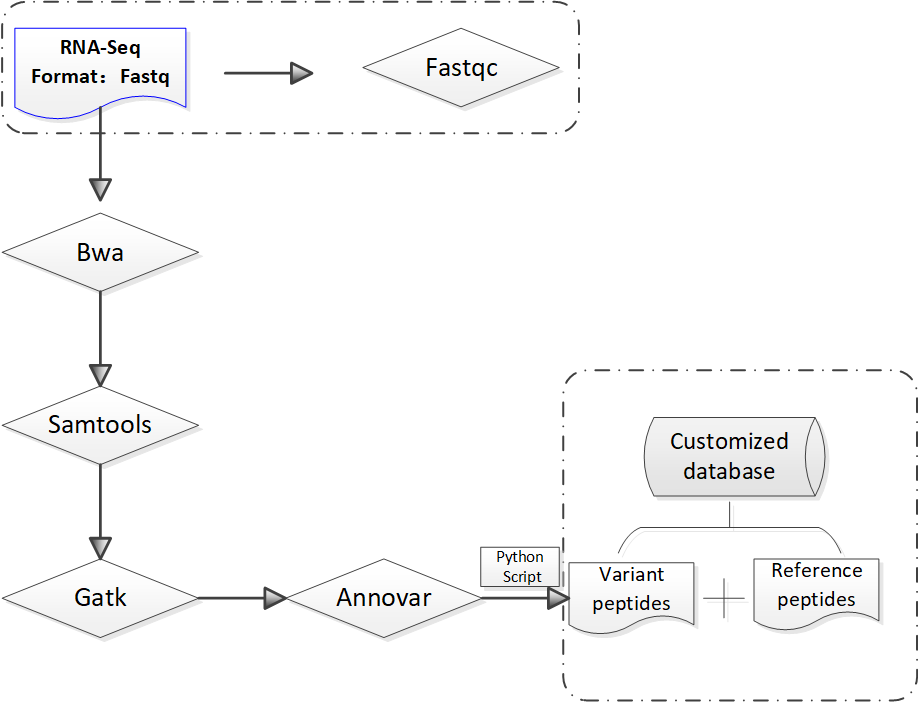
Figure1. Construction of customized protein sequence database
In order to generate the customized protein sequence database, protein sequences with missense mutation sites can be generated by substituting the mutant amino acid in normal protein sequences and all mutan sequences were appended to the normal protein and cRAP fasta file. Here we only provide mutant protein sequences (Var-proSeq.fasta) based on RNASeq data, users can add other reference protein sequences as needed.
- 1.Include samtools, razers3, hdf5 and cbc in your PATH environment variable. Add HDF5's lib directory to your LD_LIBRARY_PATH.
- 2.Installation of samtools
cd soft/samtools
./configure --prefix= /path/to/soft/
make &&make install- 3.Installation of cbc
cd soft/Cbc-2.9.9
BuildTools/get.dependencies.sh
./configure
make && make install- 4.export HDF5_DIR=/path/to/hdf5-1.8.15
- 5.install packages
pip install numpy
pip install pyomo
pip install pysam
pip install matplotlib
pip install tables
pip install pandas
pip install future- 6.Create a configuration file following config.ini In the 'OptiType' directory edit the script config.ini
cd soft/OptiType
python OptiTypePipeline.py -i /path/to/RnaSeq_1.fastq /path/to/RnaSeq_2.fastq --rna -v -o rna-hla_output eg: python OptiTypePipeline.py -i ./test/rna/CRC_81_N_1_fished.fastq ./test/rna/ CRC_81_N_2_fished.fastq --rna -v -o ./test/rna/
- 1.Installation of NetMHCpan-4.0
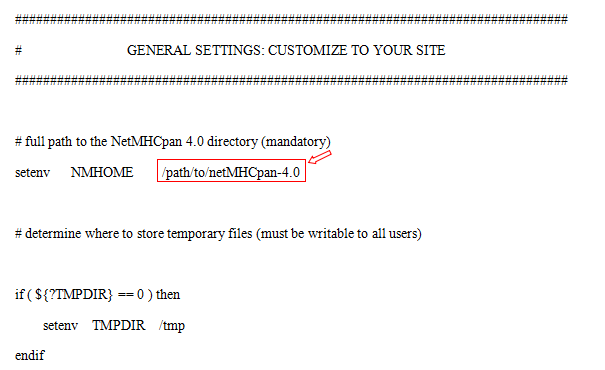
cd soft/NetMHCpan-4.0In the 'netMHCpan-4.0' directory edit the script 'netMHCpan' [7]: At the top of the file locate the part labelled "GENERAL SETTINGS: CUSTOMIZE TO YOUR SITE”, set the 'NMHOME' variable to the full path to the 'netMHCpan-4.0' directory on your system.
- 2.Installation of mono
cd soft/mono-5.18.0.225
./configure --prxfix=path/to/soft
make && make install- 3.Include netMHCpan-4.0, kallisto and blast in your PATH environment variable.
BLASTDB=~/soft/Balachandran/blast_db
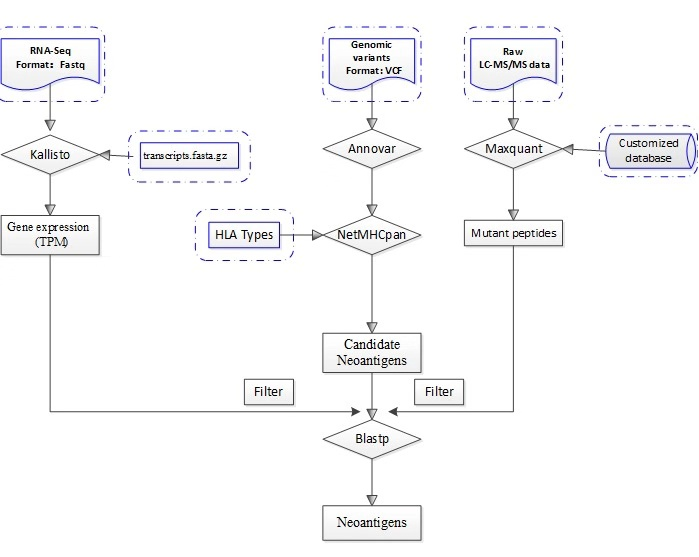
python neoantigen_prediction_filtration.py /path/to/WES.vcf HLA_typing /path/to/transcripts.fasta.gz /path/to/RnaSeq1_1.fastq /path/to/RnaSeq1_2.fastq /path/to/raw /path/to/.fasta The transcripts.fasta file supplied can be either in plaintext or gzipped format. Prebuilt indices constructed from Ensembl reference transcriptomes can be download from the kallisto transcriptome indices site [9].
eg: python NetMHCpan_Maxquant_lable-free.py test/WGS_20180423.vcf HLA-A03:01 soft/kallisto/test/transcripts.fasta.gz test/rna/rnaseq-sample1_1.fastq test/rna/rnaseq-sample1_2.fastq /export3/home/user/pipline/test/ms /export3/home/user/pipline/refseq+varseq.fasta
Figure2. Prediction and Filtration of Neontigens
Table 1 summarizes the needed software and download links
[1] Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform[M]. 2009.
[2] Li H , Handsaker B, Wysoker A , et al. The Sequence Alignment/Map format and SAMtools[J]. Bioinformatics, 2009, 25(16):2078-2079.
[3] Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–93.
[4] Ga V D A , Carneiro M , Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline.[J]. Current Protocols in Bioinformatics, 2013, 43(1110):11.10.1.
[5] Wang K , Li M , Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data[J]. Nucleic Acids Research, 2010, 38(16):e164-e164.
[6] Szolek A , Schubert B , Mohr C , et al. OptiType: precision HLA typing from next-generation sequencing data[J]. Bioinformatics, 2014, 30(23):3310-3316.
[7] Jurtz V, Paul S, Andreatta M, et al. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data[J]. Journal of Immunology, 2017, 199(9):3360.
[8] Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification[J]. Nature Biotechnology, 2008, 26(12):1367.
[9] Bray N L, Pimentel H, Melsted, Páll, et al. Near-optimal probabilistic RNA-seq quantification.[J]. Nature Biotechnology, 2016, 34(5):525.
[10] Lobo. Basic Local Alignment Search Tool (BLAST)[J]. Journal of Molecular Biology, 2012, 215(3):403-410.