Code repository of "Localized Adpative Risk Control" by Matteo Zecchin and Osvaldo Simeone.
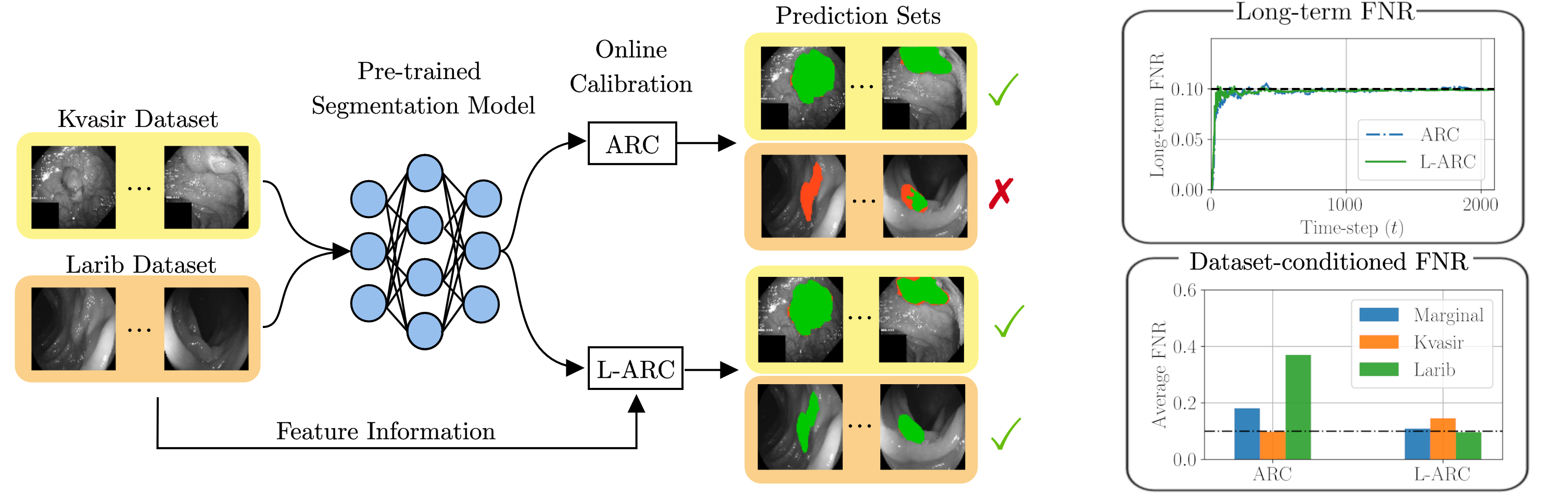 Calibration of a tumor segmentation model via ARC and the proposed localized ARC, L-ARC. Calibration data comprises images from multiple sources, namely, the Kvasir data set and the ETIS-LaribPolypDB data set. Both ARC and L-ARC achieve worst-case deterministic long-term risk control in terms of false negative rate (FNR). However, ARC does so by prioritizing Kvasir samples at the detriment of the Larib data source, for which the model has poor FNR performance. In contrast, L-ARC can yield uniformly satisfactory performance for both data subpopulations.
Calibration of a tumor segmentation model via ARC and the proposed localized ARC, L-ARC. Calibration data comprises images from multiple sources, namely, the Kvasir data set and the ETIS-LaribPolypDB data set. Both ARC and L-ARC achieve worst-case deterministic long-term risk control in terms of false negative rate (FNR). However, ARC does so by prioritizing Kvasir samples at the detriment of the Larib data source, for which the model has poor FNR performance. In contrast, L-ARC can yield uniformly satisfactory performance for both data subpopulations.
Localized Adpative Risk Control (L-ARC) is an online calibration, in which at every time set
$$
C_t=C(X_t,g_t):=\left{y\in\mathcal{Y}:s(X_t,y)\leq g_t(X_t)\right},
$$
where
Given a loss function
We provide 4 different experiments {electricity_forecast, tumor_segmentation, beam_selection, fruit_image_classification}, one in each folder.
- For the
electricity_forecastwe use the Elec2Data. To reproduce the experiment simply run themain.pyscript to run ARC and L-ARC with different localization parameters. The results will be save in the folderLogs. After that, by runningplot_figures.py, the images in the paper will be generated inside the folderImages. - For run the
tumor_segmentationexperiment we use the existing PraNet model and data. The pre-processed data, corresponding to the groudthruth masks, model's output and extracted feature can be downloaded from link. Place the.npyfiles in thecachedirectory. To produce the results of the paper, first run themain.pyscript to run ARC and L-ARC with different localization parameters and 10 different seeds. To plot the final average performance use the scriptplot_figures.py. - For the
beam selectionexperiment we include the ray-tracing files, even if they are not necessary to reproduce the experiments. These files can be modified to define different propagation scenes in Sionna. We also include the ray-tracing data in the filedataand the NN SNR predictorNN_beam_predictor. To reproduce the experiments, first run themain.pyscript. This will run ARC and L-ARC for 30 different seeds. To test the compute the average SNR over the deployment area coverage use thetest.pyscript. The final figures can then be obtained using the scriptplot_figures.pyfor the average set size and long-term coverage,plot_SNR_maps.pyfor the SNR maps andplot_SNR_thresholds.pyfor the thresholds in the appendix. - For the
fruit_image_classificationwe use the fruit-360 data and a pretrained ResNet18 model. We include the pre-processed data in the folderdata. To run the experiment, run themain.pyscript that will perform calibration using ARC and L-ARC with different localization parameters. The final results can be plotted using theplot_figures.pyscript. Even if not necessary to reproduce the experiments, in the folderResNet_TLwe include a script to perform transfer learning the ResNet18 model using the fruit-360 data.
To run the code, the following packages are necessary:
frourosto load the dataset in theelectricity_forecastexperiment.sklearnto perform PCA in thetumor_segmentationexperiment.torchto perform inference using machine learning models.sionnato generate the ray-tracing data in thebeam selectionexperiment.matplotlibto plot the figures.numpyfor array numerical operations.pickleto store and load results.