Code for the following paper:
- Xin Du, Lixin Xiu, and Kumiko Tanaka-Ishii. Bottleneck Minimal Indexing for Generative Document Retrieval. ICML 2024. Paper available at https://arxiv.org/abs/2405.10974
If you find the work useful, please cite the following paper:
@inproceedings{
du2024bottleneckminimal,
title={Bottleneck-Minimal Indexing for Generative Document Retrieval},
author={Du, Xin and Xiu, Lixin and Tanaka-Ishii, Kumiko},
booktitle={Forty-first International Conference on Machine Learning},
year={2024},
url={https://openreview.net/forum?id=MFPYCvWsNR}
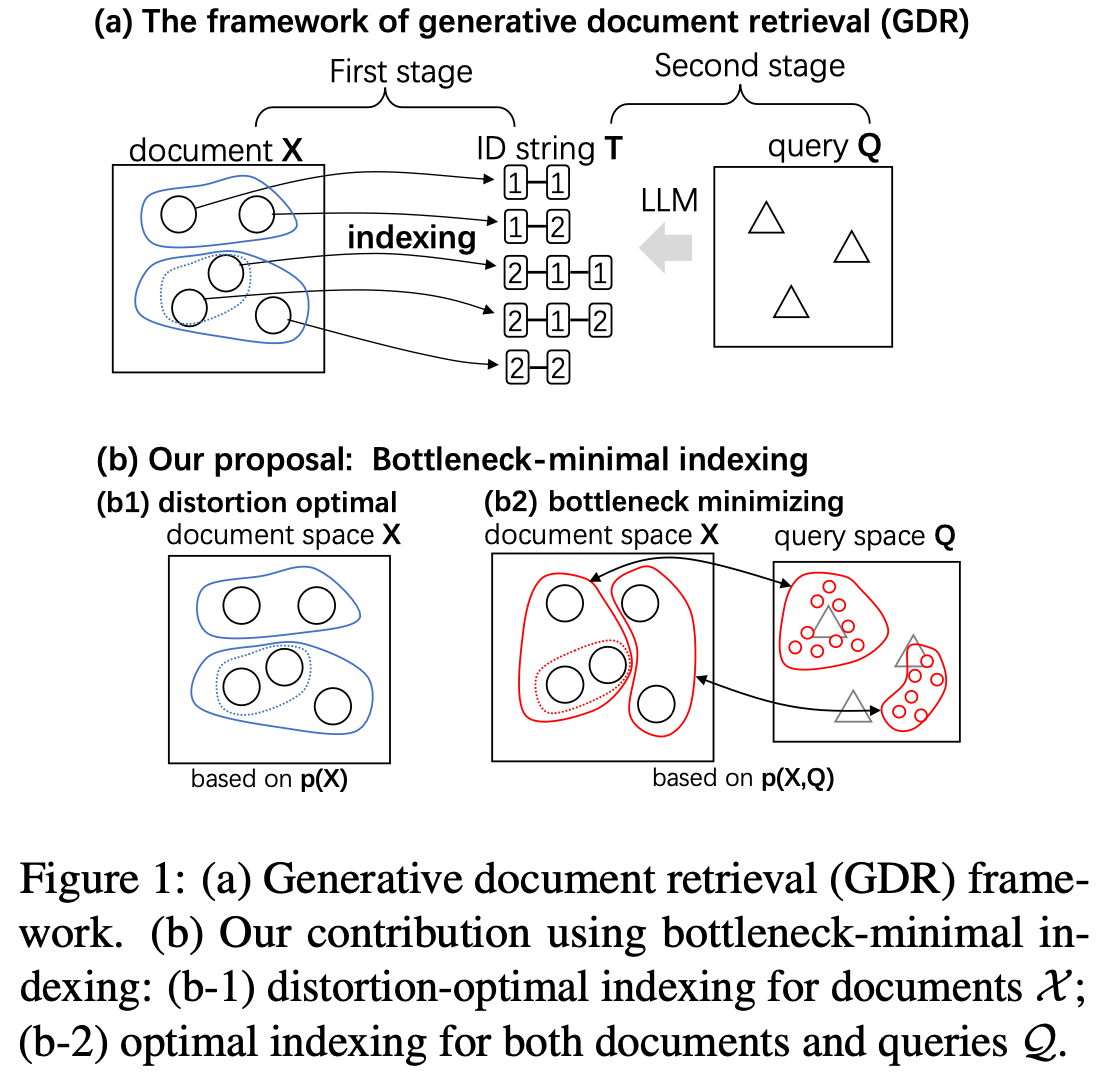
}We reconsider the document retrieval process as a data transmission problem from a query to a document, where the document's index forms an "information bottleneck." Previous works, including DSI [1] and NCI [2] for generative document retrieval (GDR), proposed indexing each document with a unique number string, such as "1-29-3-6." This indexing was produced through hierarchical k-means clustering of document embeddings. However, the optimality of k-means clustering was not theoretically explored.
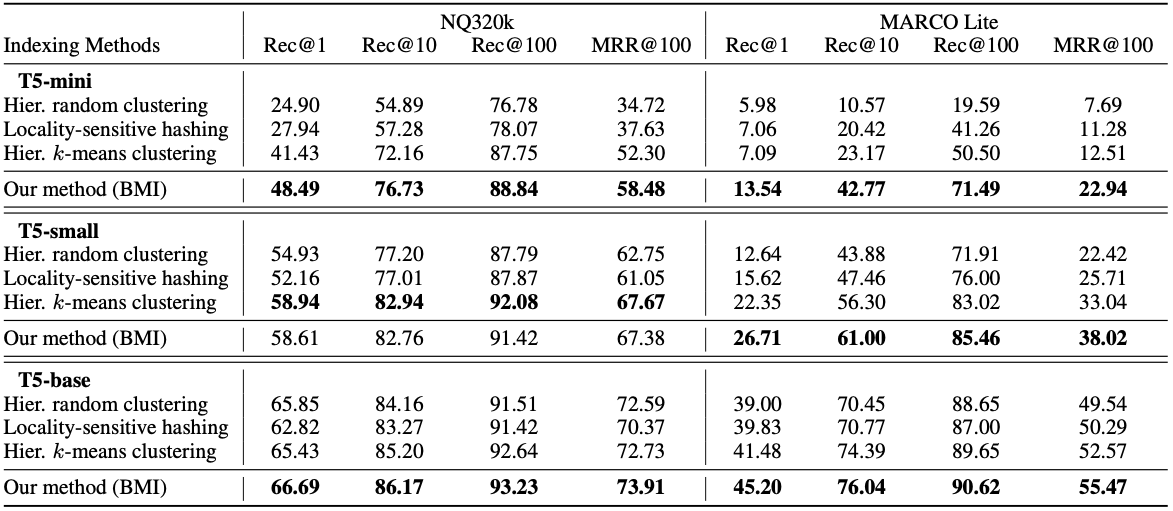
In this work, we formulate this indexing problem using the information bottleneck theory of Tishby et al. [3] and show that indexing must consider query distributions in addition to document distributions to achieve information-theoretical optimality. We propose a new indexing method called bottleneck-minimal indexing (BMI), which is a simple adaptation of the original indexing method in DSI and NCI. Instead of applying the hierarchical k-means algorithm to the document embeddings, we apply it to the query embeddings. This simple change leads to a significant improvement in retrieval performance on the NQ320K and Marco datasets, especially for smaller models, as shown in the following table.
References:
- Tay, Y., Tran, V., Dehghani, M., Ni, J., Bahri, D., Mehta, H., ... & Metzler, D. (2022). Transformer memory as a differentiable search index. Advances in Neural Information Processing Systems, 35, 21831-21843.
- Wang, Y., Hou, Y., Wang, H., Miao, Z., Wu, S., Chen, Q., ... & Yang, M. (2022). A neural corpus indexer for document retrieval. Advances in Neural Information Processing Systems, 35, 25600-25614.
- Tishby, N., Pereira, F. C., & Bialek, W. (2000). The information bottleneck method. arXiv preprint physics/0004057.
We provide the code to reproduce the results on the NQ320K dataset.
(Optionally, create a new environment with CUDA 12 support)
$ pip install -r requirements.txtPlease follow the instructions in scripts/NQ320K/preprocess_NQ320K.ipynb to
prepare the data and indexings for the original method (HKmI, Hierarchical
K-means Indexing) and our method (BMI).
To train a retrieval model based on the indexing produced by our method (BMI),
run the script in scripts/NQ320K/train_BMI.sh. Optionally, run
scripts/NQ320K/train_HKmI.sh to train a retrieval model based on the indexing
produced by the original method (HKmI).
It is recommended to use WanDB for tracking the training process. To do that, get your API key from https://wandb.ai/ and insert the key into WANDB_API_KEY in the training scripts.
Run the script in scripts/NQ320K/infer.sh. You must specify the path to the
trained model (**.ckpt) and the data in the script.
As for the MARCO-Lite dataset, please use the scripts under scripts/Marco-Lite.
To reproduce the results for T5-mini and T5-tiny, you need to download manually the two models from the Huggingface model hub. This can be done by running the following script:
$ huggingface-cli download --local-dir data/pretrained/t5-mini google/t5-efficient-mini
$ huggingface-cli download --local-dir data/pretrained/t5-tiny google/t5-efficient-tinyA large part of the code is based on the implementation of NCI. We thank the authors for their great work.