Molemap maps linked- and long-read sequences to a linear reference genome.
Individual long-reads are swiftly mapped without providing base pair precision. The mapping is multiple times faster than established long read mappers like minimap2, blend or BWA-MEM. Molemap returns mapped long reads in the SAM format.
Linked reads are mapped on the barcode level. Molemap maps all reads that share the same barcode in unity and returns a mapping file in bed format. When working on uncompressed files, a barcode index is constructed alongside the mapping which can be used to quickly retrieve all reads belonging to any given barcode. For ease of use we provide a snakemake workflow to extract all reads from user defined regions of interest.
Molemap leverages minimizers and hash tables to achieve ultra fast, memory efficient and reliable mapping of long sequence molecules. The low computational requirements of molemap allow the analysis of whole genome sequencing data of the human genome on basic hardware like a laptop.
- g++ version 7.2.0 or higher
- GNU make
- zlib dev package
Execute the following command.
git clone https://github.com/kehrlab/molemap.git
cd molemap
make
The executable 'molemap' will be created locally in the cloned repository.
- Long-reads in fastq format (can be gzip compressed)
- Tested on long-reads and HiFi-reads by PacBio and Oxford Nanopore.
- Paired-end Linked-reads in fastq format (can be gzip compressed)
- Barcodes are stored in BX:Z: flag of read Ids
- Sorted by barcode (use i.e. bcctools (only for 10x genomics linked-reads) or samtools)
- Tested on linked reads by 10x Genomics, STLfr and TellSeq
To trimm, correct and sort barcodes with bcctools use the following command in the bcctools folder:
./script/run_bcctools -f fastq <first.fq.gz> <second.fq.gz>
For detailed information on Arguments and Options:
./molemap [command] --help
Builds a minimized open addressing k-mer index of the reference genome. Linked- and long-reads require indexes with different characteristics, use the --preset option to choose the desired data type
./molemap index <reference.fa> --preset [linked/long] [options]
./molemap maplong <readfile.fq> [options]
Maps the barcodes of the provided readfiles to the reference and creates a barcode index of the readfiles to quickly retrieve all reads of a given barcode.
./molemap maplinked <readfile1.fastq> <readfile2.fastq> [options]
Content of output bed-file:
- chromosome startposition endposition barcode mapping_score
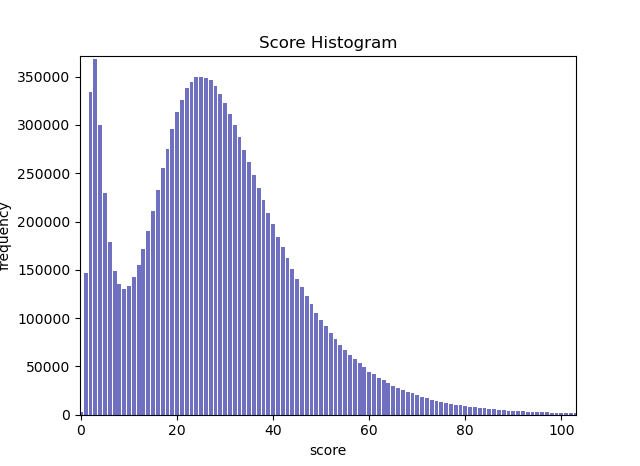
Molemap returns a output.hist file that can be ploted using plot_score_histogram.py resulting in a plot like the one above. To create a set of mappings with very high precision (at the cost of some recall), the local minimum inbetween the two peaks should be set as the score threshold. A lower theshold yields better recall at the cost of precision, a higher threshold is not recomended.
Returns all reads of the given barcodes. Barcodes can be provided directly as argument or in a file.
./molemap get <readfile1.fastq> <readfile2.fastq> <Barcodes.txt> [options]
This small example demonstrates how to use molemap and allows you to check if it is properly installed. Navigate to the molemap folder and run the commands listed below.
# building the index for chr21.fa
./molemap index example/chr21.fa -o example/Index -p linked
# mapping the reads of readfile 1 and 2 to chromosome 21
./molemap maplinked example/readfile.1.fq example/readfile.2.fq -i example/Index -r example/ReadIndex -o example/results.bed
# extracting the first barcode from the results
awk '{if(NR==1) print($4)}' example/results.bed > example/FirstBarcode.txt
# extracting all reads belonging to the first barcode
./molemap get example/readfile.1.fq example/readfile.2.fq example/FirstBarcode.txt -r example/ReadIndex -o example/readsOfFirstBarcode
# extracting reads of barcode AACATCGCAAACAGTA
./molemap get example/readfile.1.fq example/readfile.2.fq AACATCGCAAACAGTA -r example/ReadIndex -o example/readsOfAACATCGCAAACAGTA