About bs3fa
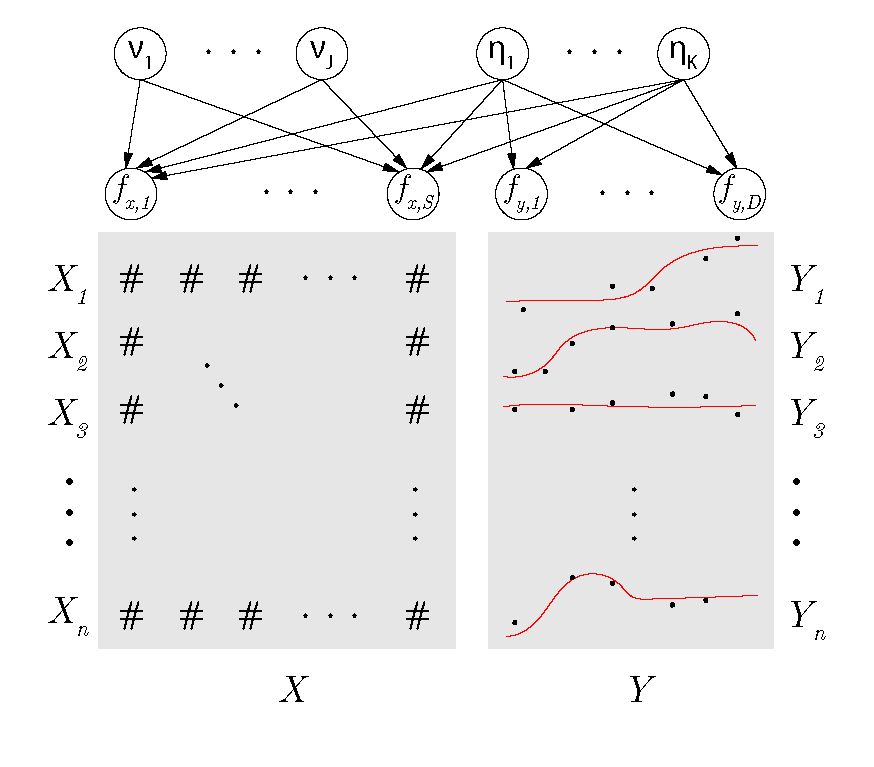
bs3fa is a package for Bayesian partially Supervised Sparse and Smooth Factor Analysis (BS3FA). This model is appropriate for data in which you have observations of functional Y and numeric (continuous, binary, count) data X. The model assumes all variation in Y is explained by some low-dimensional factors, and these factors also explain part (but not all) of the variation in X. This library can also be used when no 'supervising' X data are available and smooth unsupervised factor analysis is desired (see guide to what is contained in repository below).
Installing the package
After downloading and installing R (if necessary) and Rtools (if installing on a Windows machine) run the following from within R:
# Install packages required by bs3fa
install.packages(c("remotes", "Rcpp", "abind", "pracma", "sparseEigen",
"mvtnorm", "gridExtra", "ggplot2"))
# Install the bs3fa package
remotes::install_github("kelrenmor/bs3fa", quiet=T, upgrade=T, dependencies=T)
A workaround to the above involves downloading the repository to one's local machine and sourcing the files containing the functions provided in the package. Upon downloading the package, run the following:
setwd('/path/to/repo/bs3fa/') # set this to the full path of bs3fa
library(R.utils)
library(Rcpp)
library(RcppArmadillo)
sourceDirectory("R")
sourceCpp("src/main.cpp")
sourceCpp("src/msf.cpp")
What's in this repository
The R directory contains R functions available in this package, including run_bs3fa() (the main model sampler), run_fpca() (the sampler for Y-only smooth factor analysis), and the post-processing functions used to resolve rotational ambiguity.
The src directory contains cpp source code used for sampling specific parameters in the Gibbs sampler (i.e., this directory contains the sampling functions used within the sampler loop of run_bs3fa()) and the post-processing functions used to resolve label and sign switches.
The demo directory contains a demo of the method using realistically simulated active chemicals, and continuous chemical features with sparsity in the toxicity-relevant loadings.
The data directory contains a cleaned set of samples from the ToxCast Attagene PXR assay measured at a common grid of dose values. It is used in the simulate_data() function if the user desires more realistic loading vectors be simulated.