If you use this package in your research, please cite:
Kerseviciute I, Gordevicius J. aPEAR: an R package for autonomous visualisation of pathway enrichment networks. BioRxiv. 2023 Mar 29; doi: 10.1101/2023.03.28.534514
library(devtools)
install_github('ievaKer/aPEAR')library(devtools)
install_github('ievaKer/aPEAR@development')library(aPEAR)
library(clusterProfiler)
library(org.Hs.eg.db)
library(DOSE)
data(geneList)
enrich <- gseGO(geneList, OrgDb = org.Hs.eg.db, ont = 'CC')
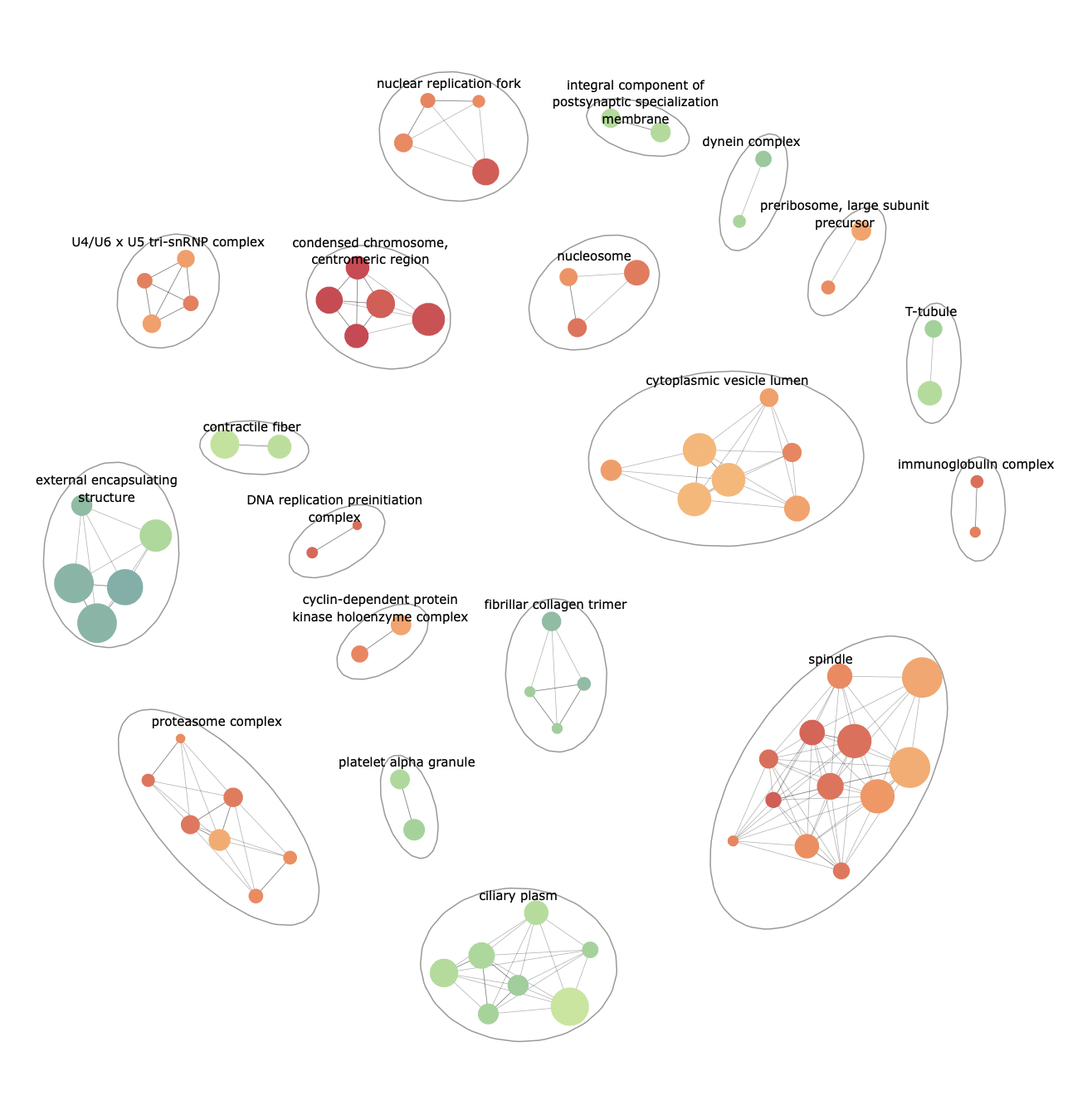
p <- enrichmentNetwork(enrich@result, drawEllipses = TRUE, fontSize = 2.5)
pTo create interactive plots, use plotly:
library(plotly)
ggplotly(p, tooltip=c('ID', 'Cluster', 'Cluster size'))