A bioinformatics pipeline for organism identification and sample-label verification of whole genome sequence (WGS) data produced at the DCLS
The DCLS is in need for a quality assurance pipeline to verify sample ID labels prior to publishing sequence data.
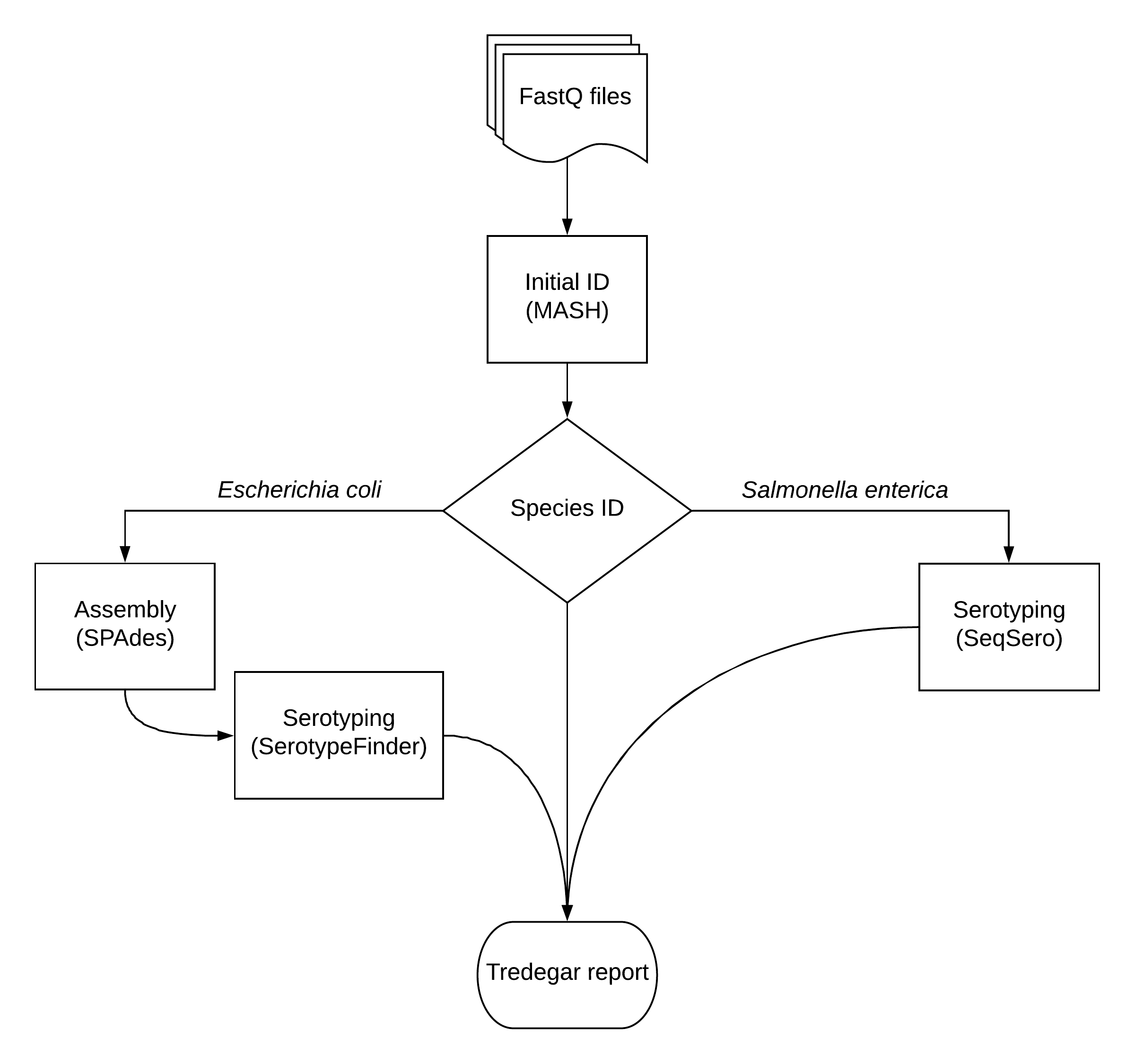
Tredegar uses MASH, a fast genome distance estimation algorithm, to determine the taxonomic identitity of all isolates; MASH is capable of accurate identification up to the species level. This level of resolution will suffice for organisms other than *Salmonella spp. and Escherichia coli isolates.
For Salmonella spp. and Escherichia coli isolates, serotype is determined through SeqSero and SerotypeFinder, respectively.
All results are compiled into a single Tredegar report. This report can be accessed by WGS scientists to compare molecular typing results of each sample; significant incongruences are assumed to be a result of mislabeling.
*The majority of isolates sequenced at the DCLS are Salonella spp. and Escherichia coli. For this reason, verification of species does little for quality assurance. E.g. if an entire sequencing run contains Salmonella enterica isoaltes of different serotypes, confirming the species will not help to identify mislabeled samples within that run.
$ tredegar.py <input_dir> -o <output_dir>
Tredegar assumes <input_dir> to be populated with paired-end fastq files; can also be a BaseMounted Project directory