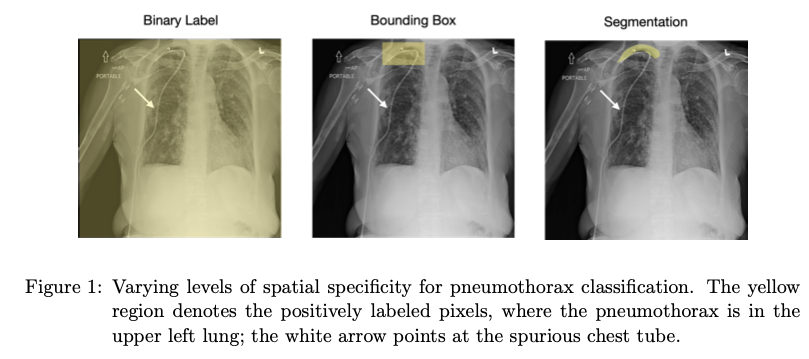
Reducing Reliance on Spurious Features with Spatial Specificity
This reposity provides implementations and experiments for the following paper:
Reducing Reliance on Spurious Features in Medical Image Classification with Spatial Specificity
Khaled Saab, Sarah Hooper, Mayee Chen, Michael Zhang, Daniel Rubin, and Christopher Ré
Machine Learning for Healthcare, 2022
Paper: https://web.stanford.edu/~ksaab/media/MLHC_2022.pdf
Requirements
Package dependencies are listed in requirements.txt.
Codebase structure
├── cfg
|=== config.yaml # default config file
├── src
├── data
|=== cxr.py # data loading code for CXR
|=== isic.py # data loading code for ISIC
|=== modeling.py # contains model architectures
|=== task.py # main lightning module
|=== utils.py
|=== cxr_tube_dict.pkl # CXR tube labels
|=== README.md
|=== requirements.txt
|=== train.py # main entry point Training
The code for training models is based on Pytorch-Lightning. Hyperparameters and config values are defined and passed using Hydra.
For example, to run the ResNet-50 ERM model:
python -m train train.method=erm train.model_type=resnet50
And to run the UNet Segmentation model:
python -m train train.method=seg train.model_type=resunet
Evaluation
After a model trains, you can load the model and evaluate its performance on different subgroups. Check out our notebook notebooks/cxr_evaluation.ipynb for an example.
Datasets
-
CXR Pneumothorax Segmentation Data
- CXR images/masks: the public SIIM-ACR Pneumothorax dataset was used, which can be found here: https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation.
- Tube labels: To evaluate robustness to the chest tube spurious feature, we hand labeled 1,951 CXRs for tubes, which we are publicly releasing. The labels can be found in the file
cxr_tube_dict.pkl, which is a dictionary where the keys are the CXR file names, and the values are the binary labels indicating the existence of a tube in the CXR.
-
ISIC Skin Lesion Data
- Images, subgroup labels, and splits can be found here: https://github.com/alceubissoto/debiasing-skin.
Citation
If you use our code, labeled data, or found our work valuable, please cite:
@article{saab2022reducing,
title={Reducing Reliance on Spurious Features in Medical Image Classification with Spatial Specificity},
author={Saab, Khaled and Hooper, Sarah and Chen, Mayee and Zhang, Michael and Rubin, Daniel and R{\'e}, Christopher},
journal={Machine Learning for Healthcare Conference},
year={2022},
organization={PMLR}
}