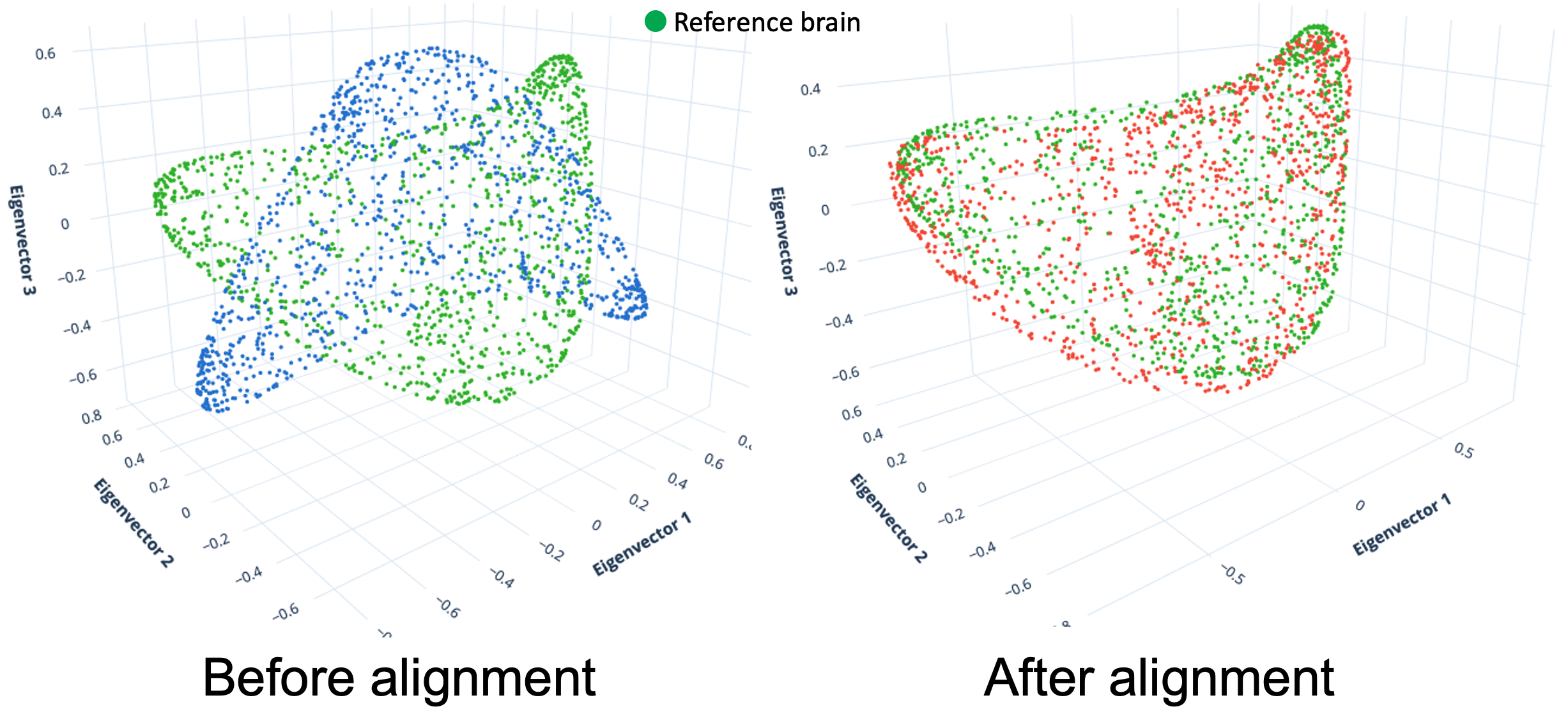
This repository impliments the python version of the graph spectral alignment.
If you find this work useful, please cite:
@article{gopinath2019graph,
title={Graph convolutions on spectral embeddings for cortical surface parcellation},
author={Gopinath, Karthik and Desrosiers, Christian and Lombaert, Herve},
journal={Medical image analysis},
volume={54},
pages={297--305},
year={2019},
publisher={Elsevier}
}
git clone https://github.com/kharitz/brain-surface-spectral-alignment
cd brain-surface-spectral-alignment
sh requirement.sh
- pytorch
- pytorch3d
- pytorch-geometric
- nibabel
- The MindBoggle brain surfaces dataset is available to download here.
- Copy the FreeSurfer directory of the input dataset to the "data" folder.
The shell script run_prep.sh will align the graph spectral of individual samples of the dataset to a reference subject in the dataset.
sh run_prep.sh
To align one sample to a reference
python spectral_align.py -r /path/to/reference/directory/
-s /path/to/sample/directory/
-o /path/to/output/directory/
--hemi lh
--eig 5
--sul
--robust
--gpu
--verbose