This repository contains a Pytorch implementation of Med3D: Transfer Learning for 3D Medical Image Analysis. Many studies have shown that the performance on deep learning is significantly affected by volume of training data. The MedicalNet project aggregated the dataset with diverse modalities, target organs, and pathologies to to build relatively large datasets. Based on this dataset, a series of 3D-ResNet pre-trained models and corresponding transfer-learning training code are provided.
MedicalNet is released under the MIT License (refer to the LICENSE file for detailso).
If you use this code or pre-trained models, please cite the following:
@article{chen2019med3d,
title={Med3D: Transfer Learning for 3D Medical Image Analysis},
author={Chen, Sihong and Ma, Kai and Zheng, Yefeng},
journal={arXiv preprint arXiv:1904.00625},
year={2019}
}
- Python 3.7.0
- PyTorch-0.4.1
- CUDA Version 9.0
- CUDNN 7.0.5
Install Python 3.7.0 pip install -r requirements.txt
- Structure of data directories
MedicalNet is used to transfer the pre-trained model to other datasets (here the MRBrainS18 dataset is used as an example).
MedicalNet/
|--datasets/:Data preprocessing module
| |--brains18.py:MRBrainS18 data preprocessing script
|--models/:Model construction module
| |--resnet.py:3D-ResNet network build script
|--utils/:tools
| |--logger.py:Logging script
|--toy_data/:For CI test
|--data/:Data storage module
| |--MRBrainS18/:MRBrainS18 dataset
| | |.../:A folder named with patient ID
| | | |reg_T1.nii.gz/:image
| | | |segm_3label_13_merge.nii.gz/:label
| |--train.txt: training data lists
| |--val.txt: validation data lists
|--pretrain/:Pre-trained models storage module
|--model.py: Network processing script
|--setting.py: Parameter setting script
|--train.py: MRBrainS18 training demo script
|--test.py: MRBrainS18 testing demo script
|--requirement.txt: Dependent library list
|--README.md
- Network structure parameter settings
Model name : parameters settings
resnet-10.pth: --model resnet --model_depth 10 --resnet_shortcut B
resnet-18.pth: --model resnet --model_depth 18 --resnet_shortcut A
resnet-34.pth: --model resnet --model_depth 34 --resnet_shortcut A
resnet-50.pth: --model resnet --model_depth 50 --resnet_shortcut B
resnet-101.pth: --model resnet --model_depth 101 --resnet_shortcut B
resnet-152.pth: --model resnet --model_depth 152 --resnet_shortcut B
resnet-200.pth: --model resnet --model_depth 200 --resnet_shortcut B
- After successfully completing basic installation, you'll be ready to run the demo.
- Clone the MedicalNet repository
git clone https://github.com/cshwhale/MedicalNet
- Downloading data & pre-trained models Unzip and move files
mv MedicalNet_pytorch_files.zip MedicalNet/.
cd MedicalNet
unzip MedicalNet_pytorch_files.zip
- Running the training code (e.g. 3D-ResNet-50)
python train.py --gpu_id 0 1 # multi-gpu training on gpu 0,1
or
python train.py --gpu_id 0 # single-gpu training on gpu 0
- Running the testing code (e.g. 3D-ResNet-50)
python test.py --gpu_id 0 --resume_path trails/models/resnet_50_epoch_200_batch_0.pth --img_list data/val.txt
- Computational Cost
GPU:NVIDIA Tesla P40
| Network | Paramerers (M) | Running time (s) |
|---|---|---|
| 3D-ResNet10 | 14.36 | 0.18 |
| 3D-ResNet18 | 32.99 | 0.19 |
| 3D-ResNet34 | 63.31 | 0.22 |
| 3D-ResNet50 | 46.21 | 0.21 |
| 3D-ResNet101 | 85.31 | 0.29 |
| 3D-ResNet152 | 117.51 | 0.34 |
| 3D-ResNet200 | 126.74 | 0.45 |
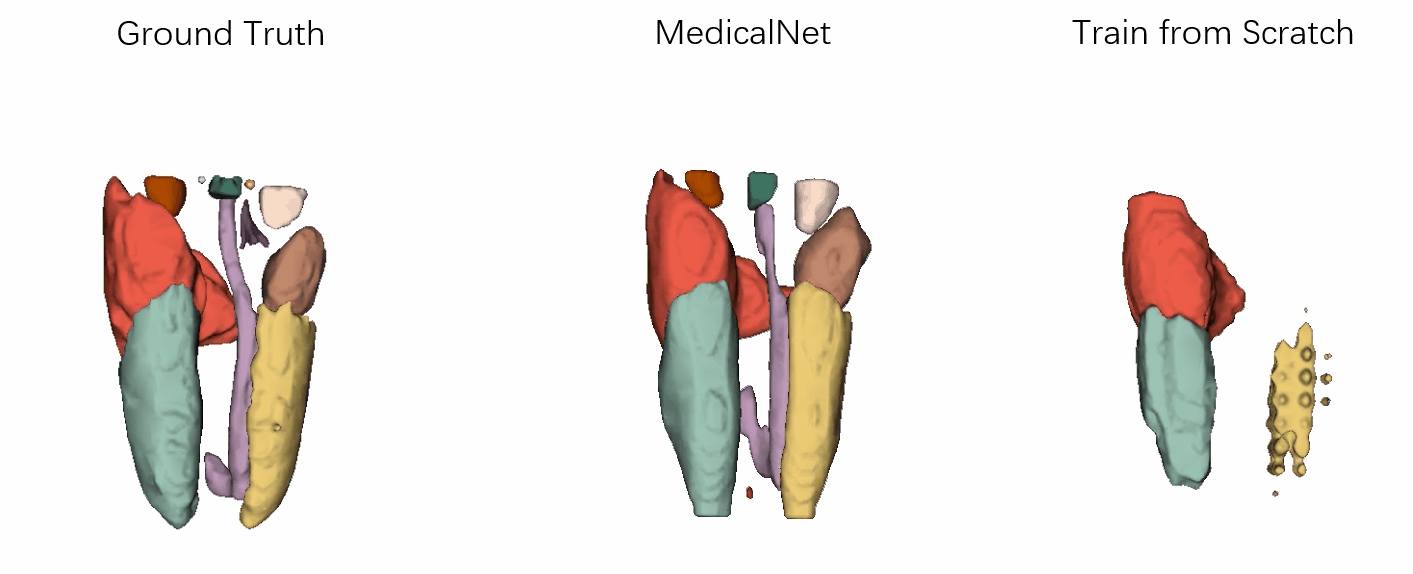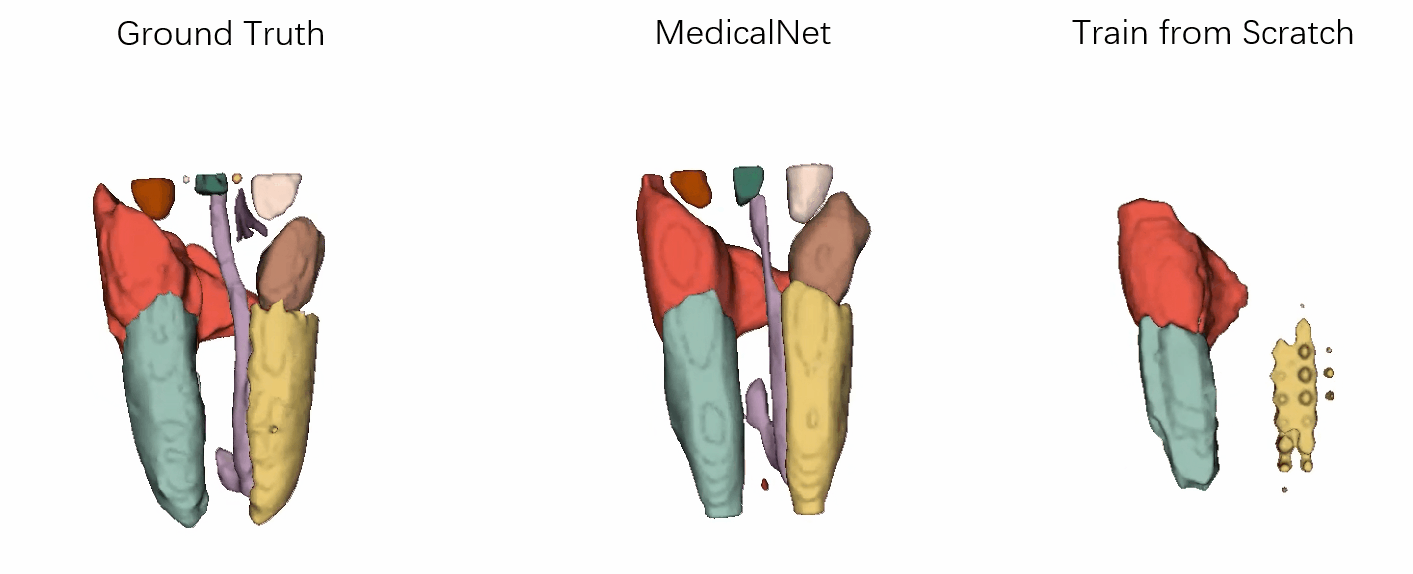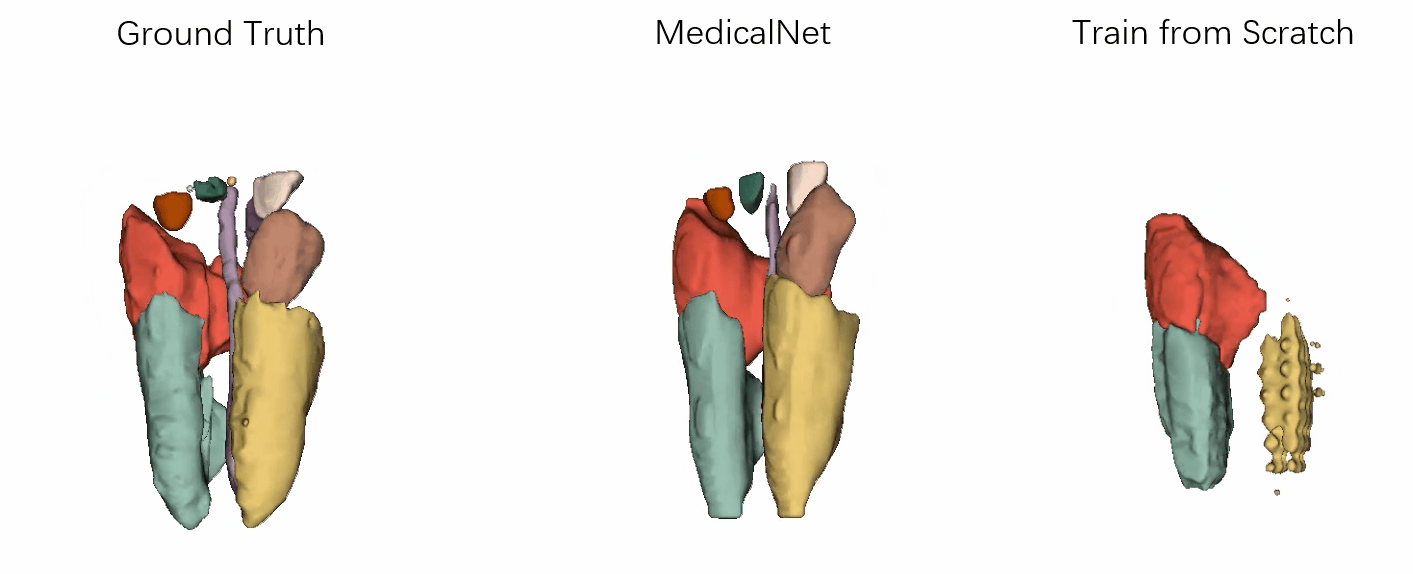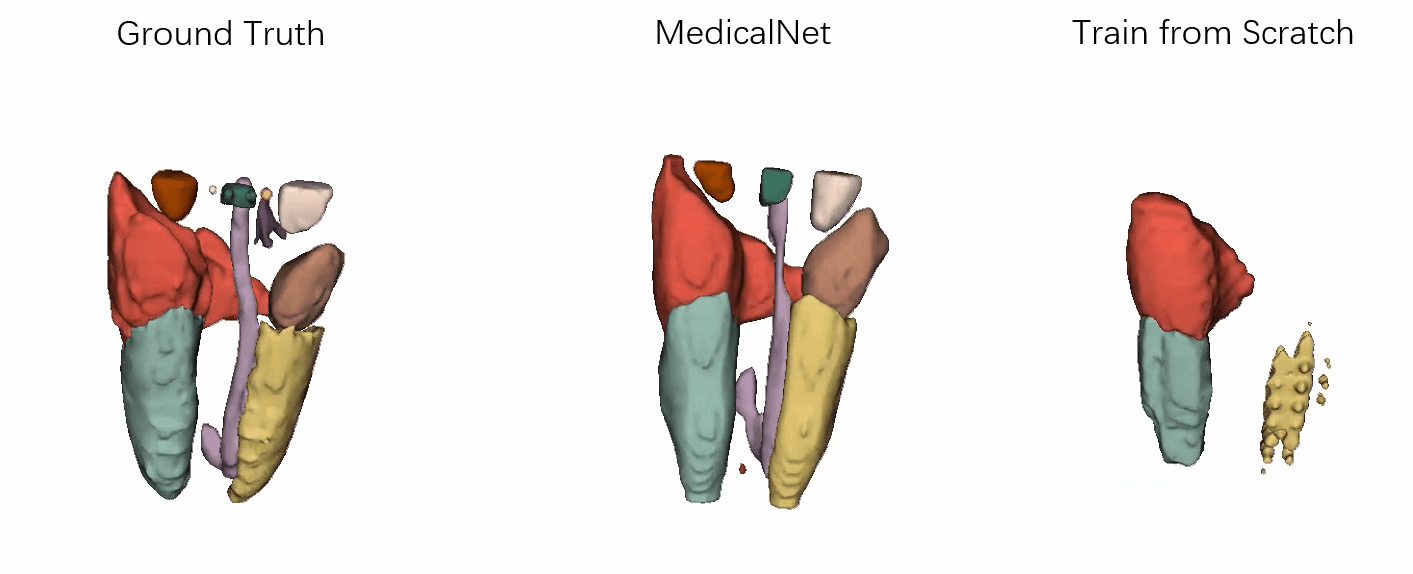
- Performance
Visualization of the segmentation results of our approach vs. the comparison ones after the same training epochs.
It has demonstrated that the efficiency for training convergence and accuracy based on our MedicalNet pre-trained models.
Results of transfer MedicalNet pre-trained models to lung segmentation (LungSeg) and pulmonary nodule classification (NoduleCls) with Dice and accuracy evaluation metrics, respectively.
| Network | Pretrain | LungSeg(Dice) | NoduleCls(accuracy) |
|---|---|---|---|
| 3D-ResNet10 | Train from scratch | 71.30% | 79.80% |
| MedicalNet | 87.16% | 86.87% | |
| 3D-ResNet18 | Train from scratch | 75.22% | 80.80% |
| MedicalNet | 87.26% | 88.89% | |
| 3D-ResNet34 | Train from scratch | 76.82% | 83.84% |
| MedicalNet | 89.31% | 89.90% | |
| 3D-ResNet50 | Train from scratch | 71.75% | 84.85% |
| MedicalNet | 93.31% | 89.90% | |
| 3D-ResNet101 | Train from scratch | 72.10% | 81.82% |
| MedicalNet | 92.79% | 90.91% | |
| 3D-ResNet152 | Train from scratch | 73.29% | 73.74% |
| MedicalNet | 92.33% | 90.91% | |
| 3D-ResNet200 | Train from scratch | 71.29% | 76.77% |
| MedicalNet | 92.06% | 90.91% |
- Please refer to Med3D: Transfer Learning for 3D Medical Image Analysis for more details:
- 3D-ResNet series pre-trained models
- Transfer learning training code
- Training with multi-gpu
- 3D efficient pre-trained models(e.g., 3D-MobileNet, 3D-ShuffleNet)
- 2D medical pre-trained models
- Pre-trained MedicalNet models based on more medical dataset
We thank 3D-ResNets-PyTorch and MRBrainS18 which we build MedicalNet refer to this releasing code and the dataset.
If you want to contribute to MedicalNet, be sure to review the contribution guidelines.