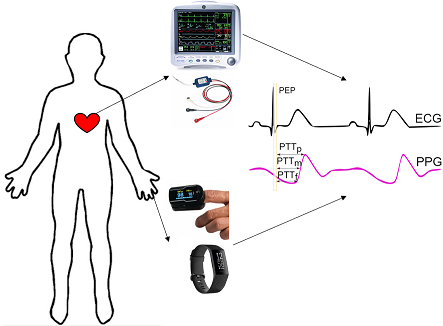
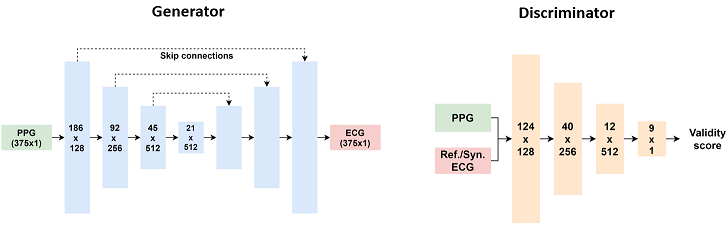
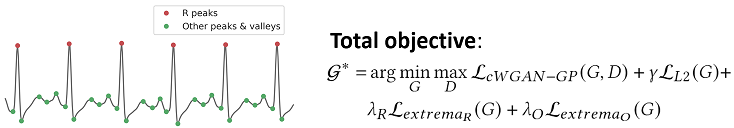Codebase for "P2E-WGAN: ECG Waveform Synthesis from PPG with Conditional Wasserstein Generative Adversarial Networks"
Paper link: https://dl.acm.org/doi/10.1145/3412841.3441979
To install the dependencies, you can run in your terminal:
pip install -r requirements.txtA sampled dataset with ECG feature indices can be downloaded at [link].
The code is structured as follows:
data.pycontains functions to transform and feed the data to the model;models.pydefines deep neural network architectures;utils.pyhas utilities to benchmark the model and calculate the gradient penalty;p2e_wgan_gp.pyis the main entry to run the training and evaluation process (support running on multiple GPUs);--dataset_prefixflag sets the directory containing the .npy files--peaks_onlyflag sets the model to reconstruct precisely only the main features for data augmentation purposes
If you find this code helpful in any way, please cite our paper:
@inproceedings{vo2021p2e,
title={P2E-WGAN: ECG waveform synthesis from PPG with conditional wasserstein generative adversarial networks},
author={Vo, Khuong and Naeini, Emad Kasaeyan and Naderi, Amir and Jilani, Daniel and Rahmani, Amir M and Dutt, Nikil and Cao, Hung},
booktitle={Proceedings of the 36th Annual ACM Symposium on Applied Computing},
pages={1030--1036},
year={2021}
}
The implementation of the WGAN-GP model is based on this repository: https://github.com/eriklindernoren/PyTorch-GAN