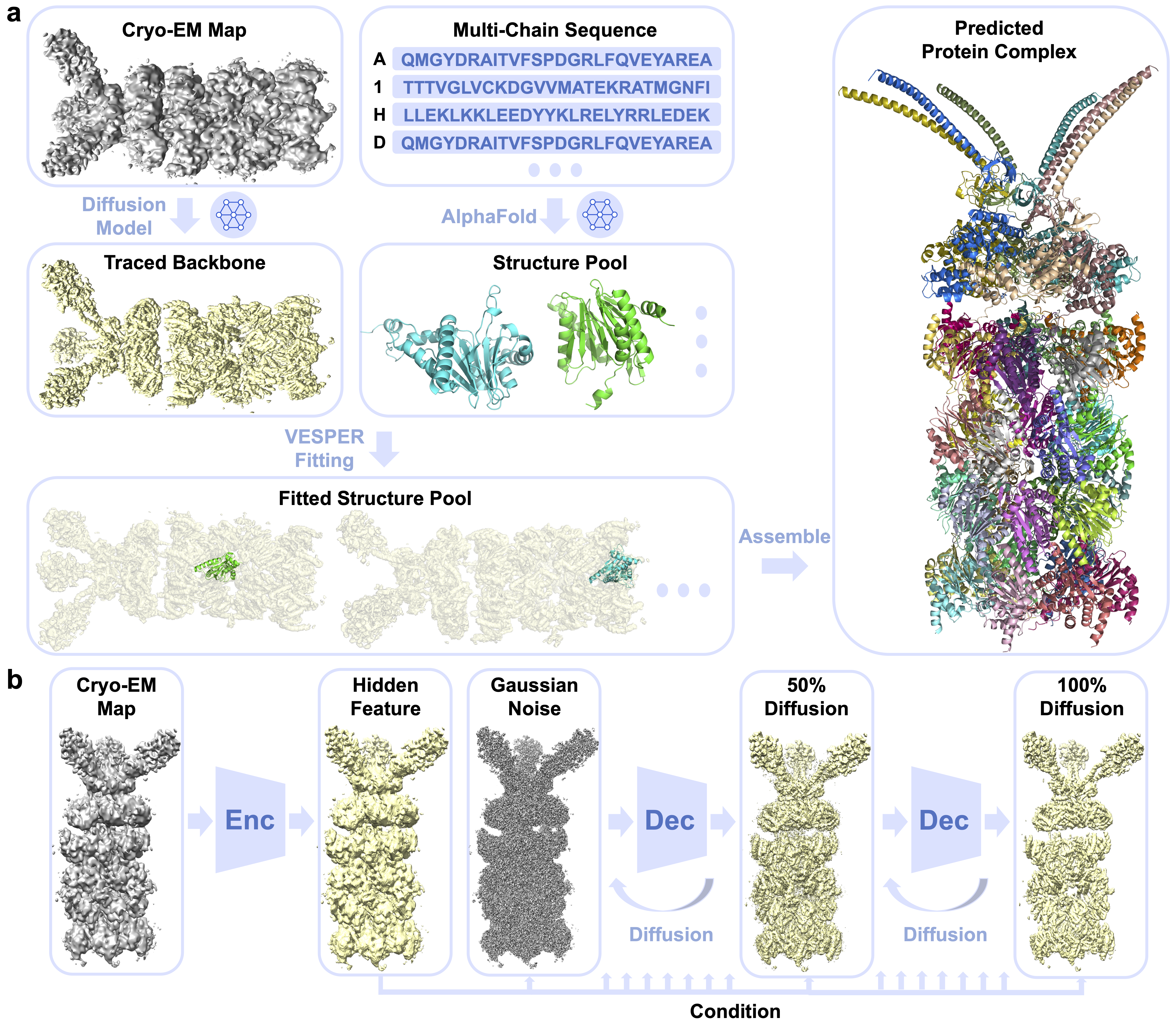
DiffModeler is a computational tool using a diffusion model to automatically build full protein complex structure from cryo-EM maps at 0-20A resolution.
Copyright (C) 2023 Xiao Wang, Han Zhu, Genki Terashi, Daisuke Kihara, and Purdue University.
License: GPL v3. (If you are interested in a different license, for example, for commercial use, please contact us.)
Contact: Daisuke Kihara (dkihara@purdue.edu)
For technical problems or questions, please reach to Xiao Wang (wang3702@purdue.edu).
The newer version of Intel MKL will cause pyTorch to give the following error due to a symbol being removed: ImportError undefined symbol: iJIT_NotifyEvent is encountered.
We have updated the environment.yml and requirements.txt to fix the version to an older version. Any previous installation should work just fine.
If you have ever encountered this issue, please activate the conda env first and run conda install mkl==2024.0.
Xiao Wang, Han Zhu, Genki Terashi & Daisuke Kihara. DiffModeler: Large Macromolecular Structure Modeling for Cryo-EM Maps Using Diffusion Model. Nature Methods, accepted 2024.
Early Version available at bioRxiv
@article{wang2023DiffModeler,
title={DiffModeler: Large Macromolecular Structure Modeling for Cryo-EM Maps Using Diffusion Model},
author={Xiao Wang, Han Zhu, Genki Terashi, Manav Taluja, and Daisuke Kihara},
journal={Nature Methods},
year={2024}
}
Input map+single-chain structures: https://em.kiharalab.org/algorithm/DiffModeler
Input map+sequence: https://em.kiharalab.org/algorithm/DiffModeler(seq)
Protein+DNA Complex structure modeling: https://em.kiharalab.org/algorithm/ComplexModeler
DiffModeler is a computational tool using a diffusion model to automatically build full protein complex structure from cryo-EM maps at 0-20A resolution.
Cryogenic electron microscopy (cryo-EM) has been widely employed in experimental settings to determine multi-chain protein complexes, but modeling accuracy greatly diminishes when resolution decreases. At intermediate resolutions of 5-10 Å, even template-based structure fitting presents significant challenges. To tackle this issue, we introduce DiffModeler, a fully automated protein complex structure modeling method that leverages a diffusion model for backbone tracing and structure fitting with AlphaFold predicted single-chain structure. In extensive testing on cryo-EM maps at intermediate resolution, DiffModeler showcased remarkably accurate structure modeling, surpassing existing methods significantly. Notably, we successfully modeled a protein complex consisting of 47 chains, comprising 13,462 residues, with an impressive TM-Score of 0.9. We also further benchmarked DiffModeler for maps at low resolution of 10-20 Å and validated its generalizability with plausible performances.
- Backbone tracing from cryo-EM maps at intermediate resolution via diffusion model.
- Single-chain structure prediction by AlphaFold.
- Single-chain structure fitting using VESPER.
- Protein complex modeling by assembling algorithms.
CPU: >=4 cores
Memory (RAM): >=12Gb.
GPU: any GPU supports CUDA with at least 12GB memory.
GPU is required for DiffModeler since most computations are done on GPU.
1. Install git
git clone git@github.com:kiharalab/DiffModeler.git && cd DiffModeler
Install anaconda from https://www.anaconda.com/download#downloads.
Then create the environment via
conda env create -f environment.yml
Each time when you want to run this software, simply activate the environment by
conda activate DiffModeler
conda deactivate(If you want to exit)
make a directory best_model and then download our pretrained model in this directory.
Diffusion model weights (trained on 5-10A, can be used for 2-5A (very good backbone tracing) and 10-20A): diffusion_model
You can also use command line to do this
mkdir best_model
cd best_model
wget https://huggingface.co/zhtronics/DiffModelerWeight/resolve/main/diffusion_best.pth.tar
cd ..
If the link failed, you can also download our model files via our lab server to best_model directory.
Pymol (for structure visualization): https://pymol.org/2/
Chimera (for map visualization): https://www.cgl.ucsf.edu/chimera/download.html
Command Parameters
usage: main.py [-h] --mode MODE [-F F] [-M M] [--config CONFIG] [--gpu GPU] [--output OUTPUT] [--contour CONTOUR]
options:
-h, --help show this help message and exit
--mode MODE control mode, mode 0: template mode; mode 1: sequence mode (online search); mode 2: sequence mode (local db search)
-F F input map path
-P P directory or zipped file of Single-Chain PDB files
-M M txt file path which records protein information
--resolution RESOLUTION
specify the resolution to skip diffusion for super high resolution maps (better than 2A)
--config CONFIG specifying the config path
--gpu GPU specify the gpu we will use
--output OUTPUT Output directory
--contour CONTOUR Contour level for input map, suggested 0.5*[author_contour]. (Float), Default value: 0.0
--fast Specify where to use fast version or not (used in server to save computations of fitting with different parameters)
--seq_search only search sequence against db, that will get templates but not do structure modeling
--af_only only search sequence against AlphaFold DB, for benchmark usage. Default: search RSCB first and then search AFDB
--domain use domain based structure for modeling, split one single-chain to multiple possible domains via SWORD2
Protein Structure Complex Modeling with provided template
This is for DiffModeler running if you have map and available template candiates (either from experimental or AlphaFold predicted structure). It is fine to run if you only know some of the template structures..
python3 main.py --mode=0 -F=[Map_Path] -P=[Single_chain_structure_dir] -M=[protin_config_path] --config=[pipeline_config_file] --contour=[Contour_Level] --gpu=[GPU_ID] --resolution=[resolution]
[Map_Path] is the path of the input experimental cryo-EM map, [Single_chain_structure_dir] specifis the directory of all single-chain PDB files. You can also zip them into a zip/.tar/.tar.gz file here to pass the zip file path here. [protin_config_path] is the text file path that records the protein single chain PDB name and corressponding chains, [pipeline_config_file] is the pipeline's parameter configuration file, saved in config directory; [Contour_Level] is the map density threshold to remove outside regions to save processing time (suggested to use half author recommended contour level), [GPU_ID] specifies the gpu used for inference. [resolution] specified the map resolution, where 0-2A will skip the diffusion model. Therefore, you can use an approximate resolution value here.
If you want to use domain-based structure modeling, you can simply add the --domain option. The program will first call SWORD2 to split each single chain into different domains and then model structures using these domain structures. Alternatively, you can use SWORD2_server to explore different domain splitting choices and provide the domain structureS as single-chain structures to DiffModeler Here DiffModeler can model protein complexes based on the domain templates you provide.
Example of PDB config file
tmp1.pdb A B
tmp2.pdb C
tmp3.pdb D E
which indicates 5 single-chain structures are provided. tmp1.pdb is for identical chain A and B, tmp2.pdb is for chain C, tmp3.pdb is for identical chain D and chain E. To obtain such template/alphafold predicted single-chain structure, please consider two options:
- Please check AlphaFold Database for single-chain structure with UniProt ID.
- Please simpy search EBI Search Tool aginst structure database to find most similar structures as templates for us to model protein complex. Here you can get similar (or even identical) experimental single-chain structures or AlphaFold predicted structures.
After running the script, the generated cif file will be kept under Predict_Result/[map_name]/DiffModeler.cif. The fitting score of each single chain is saved in occupency field of the cif file. If you want to visualize the fitting score, you can simply run the following command in PyMol after loading the cif file
spectrum q, red_white_blue, all, 0,1
Here blue indicates good fitting chains and red indicates chains may not fit well.
You can also specify --output as your output directory for your job.
python3 main.py --mode=0 -F=example/6824.mrc -P=example -M=example/input_info.txt --config=config/diffmodeler.json --contour=2 --gpu=0 --resolution=5.8
This is the example command with -P specify the directory of single-chain pdbs.
You can also use this command line to specify the zip file including all single-chain PDB files for -P
python3 main.py --mode=0 -F=example/6824.mrc -P=example/6824.zip -M=example/input_info.txt --config=config/diffmodeler.json --contour=2 --gpu=0 --resolution=5.8
Protein Structure Complex Modeling with sequence (EBI-search)
This is for DiffModeler running if you have map and corresponding sequence. It is fine to run if you only know some of the sequences.
Please use our server if with more than 4 non-identical chains. EBI's API is too slow to respond when you have many non-identical sequences.
This mode can only support structure modeling with less than 4 non-identical chains because of EBI's search tool limit.
python3 main.py --mode=1 -F=[Map_Path] -P=[fasta_path] --config=[pipeline_config_file] --contour=[Contour_Level] --gpu=[GPU_ID] --resolution=[resolution]
[Map_Path] is the path of the input experimental cryo-EM map, [fasta_path] specifis the path of sequence file with .fasta format. [pipeline_config_file] is the pipeline's parameter configuration file, saved in config directory; [Contour_Level] is the map density threshold to remove outside regions to save processing time (suggested to use half author recommended contour level), [GPU_ID] specifies the gpu used for inference. [resolution] specified the map resolution, where 0-2A will skip the diffusion model. Therefore, you can use an approximate resolution value here.
Please update email field in config/diffmodeler.json to your email.
If you want to use domain-based structure modeling, you can simply add the --domain option. The program will first call SWORD2 to split each single chain into different domains and then model structures using these domain structures. Alternatively, you can use SWORD2_server to explore different domain splitting choices and provide the domain structureS as single-chain structures to DiffModeler Here DiffModeler can model protein complexes based on the domain templates you provide.
This is based on EBI Search Tool aginst structure database to find most similar structures as templates for us to model protein complex.
Please use our server if with more than 4 non-identical chains. EBI's API is too slow to respond when you have many non-identical sequences.
Example of fasta file
>A,B,C,D
MATPAGRRASETERLLTPNPGYGTQVGTSPAPTTPTEEEDLRR
>E,F
VVTFREENTIAFRHLFLLGYSDGSDDTFAAYTQEQLYQ
For ID line, please only include the chain id without any other information. If multiple chains include the identical sequences, please use comma "," to split different chains.
In this example, we have 6 chains in total, with A,B,C,D share the identical sequences and E,F share another identical sequences.
After running the script, the generated cif file will be kept under Predict_Result/[map_name]/DiffModeler.cif. The fitting score of each single chain is saved in occupency field of the cif file. If you want to visualize the fitting score, you can simply run the following command in PyMol after loading the cif file
spectrum q, red_white_blue, all, 0,1
Here blue indicates good fitting chains and red indicates chains may not fit well.
You can also specify --output as your output directory for your job.
python3 main.py --mode=1 -F=example/6824.mrc -P=example/6824.fasta --config=config/diffmodeler.json --contour=2 --gpu=0 --resolution=5.8
Protein Structure Complex Modeling with sequence (Local Sequence Database)
This is for DiffModeler running if you have map and corresponding sequence. It is fine to run if you only know some of the sequences. If you have more 4 non-identical chains, you can set up local sequence database to run DiffModeler by yourself.
This is the mode used in server. We highly recommend to directly user server
Please follow the instructions in NCBI website to install Blast locally.
Download the processed database from https://huggingface.co/datasets/zhtronics/BLAST_RCSB_AFDB/tree/main and unzip them to data directory.
You can also use command line
wget https://huggingface.co/datasets/zhtronics/BLAST_RCSB_AFDB/resolve/main/data.tar.gz.aa
wget https://huggingface.co/datasets/zhtronics/BLAST_RCSB_AFDB/resolve/main/data.tar.gz.ab
wget https://huggingface.co/datasets/zhtronics/BLAST_RCSB_AFDB/resolve/main/data.tar.gz.ac
cat data.tar.gz.aa data.tar.gz.ab data.tar.gz.ac >data.tar.gz
tar -xzvf data.tar.gz
After configuring the environment, please run
python3 main.py --mode=2 -F=[Map_Path] -P=[fasta_path] --config=[pipeline_config_file] --contour=[Contour_Level] --gpu=[GPU_ID] --resolution=[resolution]
[Map_Path] is the path of the input experimental cryo-EM map, [fasta_path] specifis the path of sequence file with .fasta format. [pipeline_config_file] is the pipeline's parameter configuration file, saved in config directory; [Contour_Level] is the map density threshold to remove outside regions to save processing time (suggested to use half author recommended contour level), [GPU_ID] specifies the gpu used for inference. [resolution] specified the map resolution, where 0-2A will skip the diffusion model. Therefore, you can use an approximate resolution value here.
If you want to use domain-based structure modeling, you can simply add the --domain option. The program will first call SWORD2 to split each single chain into different domains and then model structures using these domain structures. Alternatively, you can use SWORD2_server to explore different domain splitting choices and provide the domain structureS as single-chain structures to DiffModeler Here DiffModeler can model protein complexes based on the domain templates you provide.
Example of fasta file
>A,B,C,D
MATPAGRRASETERLLTPNPGYGTQVGTSPAPTTPTEEEDLRR
>E,F
VVTFREENTIAFRHLFLLGYSDGSDDTFAAYTQEQLYQ
For ID line, please only include the chain id without any other information. If multiple chains include the identical sequences, please use comma "," to split different chains.
In this example, we have 6 chains in total, with A,B,C,D share the identical sequences and E,F share another identical sequences.
After running the script, the generated cif file will be kept under Predict_Result/[map_name]/DiffModeler.cif. The fitting score of each single chain is saved in occupency field of the cif file. If you want to visualize the fitting score, you can simply run the following command in PyMol after loading the cif file
spectrum q, red_white_blue, all, 0,1
Here blue indicates good fitting chains and red indicates chains may not fit well.
You can also specify --output as your output directory for your job.
python3 main.py --mode=2 -F=example/6824.mrc -P=example/6824.fasta --config=config/diffmodeler.json --contour=2 --gpu=0 --resolution=5.8
Training diffusion model in DiffModeler
To help adapt the diffusion model in DiffModeler for other purposes, we also released the training script and instructions here.
The dataset should be prepared in a directory [data_path] (keep in mind that you will use later), where the directory organization should be organized as
-[EMD-ID1]
--input_1.npy
--output_1.npy
--input_2.npy
--output_2.npy
...
-[EMD-ID2]
--input_1.npy
--output_1.npy
--input_2.npy
--output_2.npy
...
...
Here each sub-directory is the training input/target pairs collected from different EM mpas. The input_[k].npy and output_[k].npy corresponds to kth exampls's input and target. Their shape should be [K,W,H], K indicates the number of channels, W refers to the width of the box, H refers to the height of the box.
The map ids [EMD-ID] used for training and validation should be prepared in a txt file, with each line records one [EMD-ID]. This record txt file should be saved in [info_txt_path] (keep in mind that you will use later).
Depends on which channel of K channels you will use in the prepared file, you can specify them in config/diffmodeler_train.json.
"data": {
"input_channel": [0],
"output_channel":[0] //to support multi-channel training examples you have
},
This is a list configuration for both input and output, you can add many channels as you want. If you changed to more channels, please also update network configuration accordingly.
"unet": {
"in_channel": 3,
"out_channel": 1,
...
After proper configuration, you can then train your model with the following scripts:
python3 train.py -F [data_path] --info_txt [info_txt_path] --config config/diffmodeler_train.json --gpu [gpu_id] --output [output_path]
[data_path] and [info_txt_path] is the path that you configured the training data path and records path.
[gpu_id] specifies the GPU used for training diffusion model.
[output_path] is the path that you specified to save the training log and models.
If you wanted to change other configurations for better training, you can modify optimizer, network architecture, learning rate etc. in the configuration json file: config/diffmodeler_train.json
Evaluate modeled structure by DiffModeler
First convert .cif format to .pdb format via maxit: install maxit
Then run the following command to convert DiffModeler.cif to DiffModeler.pdb:
maxit -input DiffModeler.cif -output DiffModeler.pdb -o 2
For big structure that with more than 9999 residues, please reindex residue id from 1 for each chain. Otherwise, the conversion may lead to incorrect evaluation results.
You can choose to install MMAlign or run it online: MMalign.
Then run the following command to compare DiffModeler.pdb with native.pdb.
./MMalign DiffModeler.pdb native.pdb >report.txt
The evaluation metrics are available in report.txt.
The TM-score is the 2nd one, which is normalized by the 2nd structure(native.pdb).
The Align Ratio is calculated by dividing the reported align length by the length of the native structure.
The sequence identity is calculated by Seq_ID*Align-Ratio.
Cryo-EM map with mrc format. AlphaFold/Template single-chain structure and information file to indicate the path. Our example input can be found here
DiffModeler.cif: a CIF file that records the final modeled protein complex structure. Our example output can be found here. All the intermediate results are also kept here.