This repository contains the source of MolPAL, a software for the accelerated discovery of compounds in high-throughput virtual screening environments, as originally detailed in the paper Accelerating high-throughput virtual screening through molecular pool-based active learning
- Overview
- Table of Contents
- Requirements
- Installation
- Object Model
- Preprocessing
- Running MolPAL
- Future Directions
- Reproducing Experimental Results
- Python (>= 3.6)
if utilizing GPU accelerated model training/inference
- CUDA (>= 10.2)
if performing docking online
- the appropriate requirements as listed in the
pyscreenerREADME
The first step in installing MolPAL is to clone this repository: git clone <this_repo>
The easiest way to install all dependencies is to use conda along with the supplied environment.yml file, but you may also install them manually, if desired. All libraries listed in that file are required before using MolPAL
The following packages are optional to install before running MolPAL:
- cudatoolkit (whichever version matches your CUDA build if utilizing GPU acceleration for PyTorch-based models (MPN)
- map4 and tmap (if utilizing the map4 fingerprint)
- optuna (if planning to perform hyperparameter optimization)
NOTE: you may wish to edit environment.yml to reflect the CUDA version you will be using. It is currently configured to use CUDA 11.1, but you can delete this line and uncomment the other line if your setup required CUDA 10.2. If you need a lower CUDA version than that, please go to the pytorch wesbite to set the channels and versions of both pytorch and other packages properly.
- (if necessary) install conda
cd /path/to/molpalconda env create -f environment.yml
Before running MolPAL, be sure to first activate the environment: conda activate molpal
MolPAL parallelizes objective function calculation and model inference (training coming later) using the ray library. MolPAL will automatically start a ray cluster if none exists, but this is highly limiting because it can't leverage distributed resources nor will it accurately reflect allocated resources (i.e, it will think you have access to all N cores on a cluster node, regardless of your allocation.)
To properly leverage multi-node allocations, you must set up a ray cluster manually before running MolPAL. The documentation has several examples of how to set up a ray cluster, and the only thing specific to MolPAL is the reliance on two environment variables: redis_password and ip_head. MolPAL will use the values of these environment variables to connect to the proper ray cluster. An example of this may be seen in the SLURM submission script run_molpal.batch
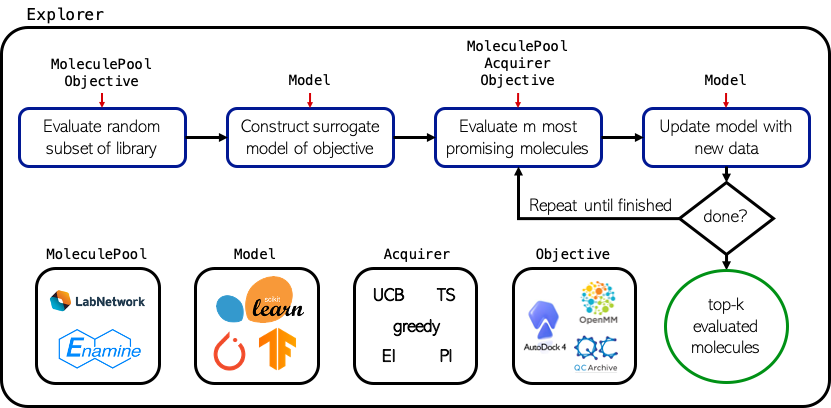
MolPAL is a software for batched, Bayesian optimization in a virtual screening environment. At the core of this software is the molpal library, which implements several classes that handle specific elements of the optimization routine.
Explorer: An Explorer is the abstraction of the optimization routine. It ties together the MoleculePool, Acquirer, Encoder, Model, and Objective, which each handle (roughly) a single step of a Bayesian optimization loop, into a full optimization procedure. Its main functionality is defined by the run() method, which performs the optimization until a stopping condition is met, but it also defines other convenience functions that make it amenable to running a single iteration of the optimization loop and interrogating its current state if optimization is desired to be run interactively.
MoleculePool: A MoleculePool defines the virtual library (i.e., domain of inputs) and caches precomputed feature representations, if feasible.
Acquirer: An Acquirer handles acquisition of unlabeled inputs from the MoleculePool according to its metric and the prior distribution over the data. The metric is a function that takes an input array of predictions and returns an array of equal dimension containing acquisition utilities.
Featurizer: A Featurizer computes the uncompressed feature representation of an input based on its identifier for use with clustering and models that expect vectors as inputs.
Model: A Model is trained on labeled data to produce a posterior distribution that guides the sequential round of acquisition
Objective: An Objective handles calculation of the objective function for unlabeled inputs
For models expecting vectors as inputs (e.g., random forest and feed-forward neural network models,) molecular fingerprints must be calculated first. Given that the set of fingerprints used for inference is the same each time, it makes sense to cache these fingerprints, and that's exactly what the base MoleculePool (also referred to as an EagerMoleculePool) does. However, the complete set of fingerprints for most libraries would be too large to cache entirely in memory on most systems, so we instead store them on disk in an HDF5 file that is transparently prepared for the user during MolPAL startup (if not already provided with the --fps option.)
If you wish to prepare this file ahead of time, you can use scripts/fingerprints.py to do just this. While this process can be parallelized over an infinitely large ray cluster (see above,) in our tests we were I/O limited above 12 cores, which takes about 4 hours to prepare an HDF5 file of 100M fingerprints. Note: if MolPAL prepares the file for you, it prints a message saying where the file was written to (usually under the $TMP directory) and whether there were invalid SMILES. To reuse this fingerprints file, simply move this file to a persistent directory after MolPAL has completed its run. Additionally, if there were no invalid smiles, you can pass the --validated flag in the options to further speed up MolPAL startup.
To prepare the fingerprints file corresopnding to the sample command below, issue the following command: python scripts/fingerprints.py --library libraries/Enamine50k.csv.gz --fingerprint pair --length 2048 --radius 2 --name libraries/fps_enamine50k
The resulting fingerprint file will be located in your current working directory as libraries/fps_enamine50k.h5. To use this in the sample command below, add --fps libraries/fps_enamine50k.h5 to the argument list.
The general command to run MolPAL is as follows:
python molpal.py -o <objective_type> [additional objective arguments] --libary <path/to/library.csv[.gz]> [additional library arguments] [additional model/encoding/acquistion/stopping/logging arguments]
Alternatively, you may use a configuration file to run MolPAL, like so:
python molpal.py --config <path/to/config_file>
Two sample configuration files are provided: minimal_config.ini, a configuration file specifying only the necessary arguments to run MolPAL, and sample_config.ini, a configuration file containing a few common options to specify (but not all possible options.)
Configuration files accept the following syntaxes:
--arg value(argparse)arg: value(YAML)arg = value(INI)arg value
A sample command to run one of the experiments used to generate data in the initial publication is as follows:
python run.py --config config_expts/Enamine50k_retrain.ini --name molpal_50k --metric greedy --init-size 0.01 --batch-size 0.01 --model rf
or the full command:
python run.py --name molpal_50k --write-intermediate --write-final --retrain-from-scratch --library libraries/Enamine50k.csv.gz --validated --metric greedy --init-size 0.01 --batch-size 0.01 --model rf --fingerprint pair --length 2048 --radius 2 --objective lookup --lookup-path data/Enamine50k_scores.csv.gz --lookup-smiles-col 0 --lookup-data-col 1 --minimize --top-k 0.01 --window-size 10 --delta 0.01 --max-epochs 5
The primary purpose of MolPAL is to accelerate virtual screens in a prospective manner. Currently (December 2020), MolPAL supports computational docking screens using the pyscreener library
-o or --objective: The objective function you would like to use. Choices include docking for docking objectives and lookup for lookup objectives. There are additional arguments for each type of objective.
docking: given the variety of screening options allowed by thepyscreenerlibrary, it's likely easiest to specify an--objective-configrather than providing these options on the command line. Theobjective-configfile must be provided in the format of apyscreenerconfiguration file, so some options might have different names (e.g.,sizein that file rather than--box-size). Any options specified on the command line will override any options provided in the configuration file.--software: the docking software you would like to use. Choices: 'vina', 'smina', 'psovina', 'qvina', and 'ucsfdock' (Default = 'vina').--receptor': the filepath of the receptor you are attempting to dock ligands into.--box-center: the x-, y-, and z-coordinates (Å) of the center of the docking box.--box-size: the x-, y-, and z- radii of the docking box in Å.--docked-ligand-file: the name of a file containing the coordinates of a docked/bound ligand. If using Vina-type software, this file must be a PDB format file. Either--box-centerand--box-sizemust be specified or a docked ligand file must be provided. In the case that both are provided,--score-mode: the method by which to calculate an overall score from multiple scored conformations
lookup--lookup-path: the filepath of a CSV file containing score information for each input
--library: the filepath of a CSV file containing the virtual library as SMILES strings
- (optional)
--fps: the filepath of an hdf5 file containing the precomputed fingerprints of your virtual library. MolPAL relies on the assumption that the ordering of the fingerprints in this file is exactly the same as that of the library file and that the encoder used to generate these fingerprints is exactly the same as the one used for model training. MolPAL handles writing this file for you if unspecified, so this option is mostly useful for avoiding the overhead at startup of running MolPAL again with the same library/encoder settings.
MolPAL has a number of different model architectures, encodings, acquisition metrics, and stopping criteria to choose from. Many of these choices have default settings that were arrived at through hyperparameter optimization, but your circumstances may call for modifying these choices. To see the full list, run MolPAL with either the -h or --help flags. A few common options to specify are shown below.
-k: the fraction (if between 0 and 1) or number (if greather than 1) of top scores to evaluate when calculating an average. (Default = 0.005)
--window-size and --delta: the principle stopping criterion of MolPAL is whether or not the current top-k average score is better than the moving average of the window_size most recent top-k average scores by at least delta. (Default: window_size = 3, delta = 0.1)
--max-explore: if you would like to limit MolPAL to exploring a fixed fraction of the libary or number of inputs, you can specify that by setting this value. (Default = 1.0)
--max-epochs: Alternatively, you may specify the maximum number of epochs of exploration. (Default = 50)
--model: the type of model to use. Choices include rf, gp, nn, and mpn. (Default = rf)
--conf-method: the confidence estimation method to use for the NN or MPN models. Choices includeensemble,dropout,mve, andnone. (Default = 'none'). NOTE: the MPN model does not support ensembling
--metric: the acquisition metric to use. Choices include random, greedy, ucb, pi, ei, thompson, and threshold (Default = greedy.) Some metrics include additional settings (e.g. the β value for ucb.)
MolPAL will automatically use a GPU if it detects one. If this is undesired, use the following command before running: export CUDA_VISIBLE_DEVICES=''
While the default settings of MolPAL were chosen based on hyperparameter optimization with Optuna, they were calculated based on the context of structure-based discovery our computational resources. It is possible that these settings are not optimal for your particular problem. To adapt MolPAL to new circumstances, we recommend first generating a dataset that is representative of your particular problem then peforming hyperparameter optimization of your own using the LookupObjective class. This class acts as an Oracle for your particular objective function, enabling both consistent and near-instant calculation of the objective function for a particular input, saving time during hyperparameter optimization.
Though MolPAL was originally intended for use with protein-ligand docking screens, it was designed with modularity in mind and is easily extendable to other settings as well. In principle, all that is required to adapt MolPAL to a new problem is to write a custom Objective subclass that implements the calc method. This method takes a sequence SMILES strings as an input and returns a mapping from SMILES string -> objective function value to be utilized by the Explorer. To this end, we are currently exploring the extension of MolPAL to subsequent stages of virtual discovery (MD, DFT, etc.) If you make use of the MolPAL library by implementing a new Objective subclass, we would be happy to include your work in the main branch.
The data used in the original publication was generated through the corresponding configuration files located in config_experiments and the library name (e.g., '10k', '50k', 'HTS', or 'AmpC') as the two command line arguments. The submission script was designed to be used with a SLURM scheduler, but if you want to rerun the experiemnts on your machine, then you can simply follow the submission script logic to generate the proper command line arguments or write a new configuration file. The AmpC data was too large to include in this repo, but it may be downloaded from here.
Once all of the data were generated, the directories were containing the data from each run organized according to the following structure:
<library> (= `--parent-dir`)
├── online
│ ├── <batch_size>
| | ├── <model>
| | | ├── <metric>
| | | | └── rep_<i>
│ │ | └── ...
│ │ └── ...
│ ├── <batch_size>
| | ├── <model>
│ │ | └── ...
│ │ └── ...
| └── ...
└── retrain
├── <batch_size>
│ └── ...
└── ...
where everything between angled brackets is a single word that describes the corresponding parameter (e.g., <model> = mpn) and <i> is a number signifiying which repeat that run represents. Each rep_<i> folder is the actual output folder from a run of MolPAL.
After the data was organized as above, scripts/analyze_data.py was used to analyze the data by parsing the output from individual runs, organizing the data, and using the data to produce the corresponding figure.
To reproduce the 10k and 50k figures in the main text, we ran the script with the following command: python scripts/analyze_data.py --parent-dir molpal_results/10k --true-pkl <10k_score_dict.pkl> --mode model-metrics --split 0.010. In our case, the full results of all the 10k runs were stored under the folder molpal_results/10k according to the directory structure above. Also note that <10k_score_dict.pkl> is a pickled python dictionary generated by scripts/make_dict.py (just a dictionary of the included score CSV files.)
For the remaining figures (e.g., UMAP and histogram figures,) use the corresponding scripts in the scripts directory. To figure out how to run them, use the following command python <script>.py --help.
the following timings used Intel Xeon 6230 CPUs and Nvidia GeForce RTX 2080 TI GPUs
| action | resources | approximate time |
|---|---|---|
| calculating 2M fingerprints | 8 CPU 1 | 4m |
| calculating 100M fingerprints | 12 CPU | 4h |
| MPN training on 2k molecules | 8 CPU / 1 GPU | 2s / epoch |
| MPN prediction on 2M molecules | 8 CPU / 1 GPU | 15m |
| MPN training on 100k molecules | 12 CPU / 1 GPU 2 | 30s / epoch |
| MPN prediction on 100M molecules | 4 x (12 CPU / 1 GPU) 2 | 2h |
1fingerprint code currently only support single process writing to limit total memory footprint. We have found it is I/O limited beyond 8 CPUs
2used Intel Xeon 6130 CPUs and Nvidia V100 GPUs