spoon is a method to address the mean-variance relationship in
spatially resolved transcriptomics data. Current approaches rank
spatially variable genes based on either p-values or some effect size,
such as the proportion of spatially variable genes. However, previous
work in RNA-sequencing has shown that a technical bias, referred to as
the "mean-variance relationship", exists in these data in that the
gene-level variance is correlated with mean RNA expression. We found
that there is a "mean-variance relationship" in spatial transcriptomics
data, and so we propose spoon, a statistical framework to prioritize
spatially variable genes that is not confounded by this relationship. We
fit a spline curve to estimate the mean-variance relationship. Then,
similar to using weights in a weighted least squares model, we used
weights that we plugged into a Gaussian Process Regression model fit
with a nearest-neighbor Gaussian process model to the preprocessed
expression measurements for each gene, i.e. one model per gene. spoon
removes the bias and leads to a more informative set of spatially
variable genes.
The generate_weights() function calculates individual observation
weights, where an individual observation is a UMI (unique molecular
identifier) count value for a specific gene and sample. If the desired
SVG detection method accepts weights, then the individual observation
weights can be used as inputs. If the desired SVG detection method does
not accept weights, then the Delta method is leveraged to rescale the
data and covariates by the weights. These scaled data and covariates are
used as inputs into the desired SVG detection function.
Bioconductor houses the infrastructure to store and analyze spatially
resolved transcriptomics data for R users, including many SVG detection
methods. This method addresses the mean-variance relationship
confounding SVG detection, which is related to these other Bioconductor
packages. Additionally, spoon is inspired by limma::voom() , which
is a popular Bioconductor package.
The following code will install the latest release version of the
spoon package from Bioconductor. Additional details are shown on the
Bioconductor page.
install.packages("BiocManager")
BiocManager::install("spoon")
The latest development version can also be installed from the devel
version of Bioconductor or from
GitHub.
We recommend the input data be provided as a
SpatialExperiment
Bioconductor object. The outputs are stored in the rowData of the
SpatialExperiment object. The examples below use this input data
format.
The inputs can also be provided as a numeric matrix of raw counts and a numeric matrix of spatial coordinates.
Load packages and data
library(nnSVG)
library(STexampleData)
library(SpatialExperiment)
library(BRISC)
library(BiocParallel)
library(scuttle)
library(Matrix)
library(spoon)
spe <- Visium_mouseCoronal()
Preprocessing
# keep spots over tissue
spe <- spe[, colData(spe)$in_tissue == 1]
# filter out low quality genes
spe <- filter_genes(spe)
# calculate logcounts (log-transformed normalized counts) using scran package
spe <- computeLibraryFactors(spe)
spe <- logNormCounts(spe)
# choose a small number of genes for this example to run quickly
set.seed(3)
ix_random <- sample(seq_len(nrow(spe)), 10)
spe <- spe[ix_random, ]
# remove spots with zero counts
spe <- spe[, colSums(logcounts(spe)) > 0]
Step 1: generate weights
weights <- generate_weights(input = spe,
stabilize = TRUE,
BPPARAM = MulticoreParam(workers = 1,
RNGseed = 4))
Step 2: weighted SVG detection
spe <- weighted_nnSVG(input = spe,
w = weights,
BPPARAM = MulticoreParam(workers = 1, RNGseed = 5))
Show results
# display results
rowData(spe)
Other SVG detection tools
We provided a function to use the weights with nnSVG for more accurate spatially variable gene detection. The weights can also be used with other spatially variable gene detection tools using the following procedure:
assay_name <- "logcounts"
weighted_logcounts <- t(weights)*assays(spe)[[assay_name]]
assay(spe, "weighted_logcounts") <- weighted_logcounts
weighted_logcounts can be accessed from
assay(spe, "weighted_logcounts"). Then, weighted_logcounts should be
used as the input counts matrix and weights as the input covariate
matrix in a spatially variable detection tool.
In the Tutorial section, we showed how to use the functions in spoon
on a small number of genes for a faster runtime. This section will show
how these methods address the mean-variance relationship in spatial
transcriptomics data. The code below takes several hours to run, but can
be reproduced if desired.
Simulate data
library(SpatialExperiment)
library(STexampleData)
library(MASS)
library(scuttle)
set.seed(1)
#4992 spots and 300 genes
n_genes <- 300
fraction <- 0.5
max_sigma.sq <- 1
low_range_beta <- c(0.5,1)
#check if integer
stopifnot(n_genes*fraction*0.5 == round(n_genes*fraction*0.5))
#all genes have some nonzero sigma.sq
sigma.sq <- runif(n_genes, 0.2, max_sigma.sq)
ground_truth_rank <- rank(-sigma.sq)
#all genes will have nonzero beta values
beta <- runif(n_genes, log(low_range_beta[1]), log(low_range_beta[2]))
#choose fixed length scale parameter (~medium from nnSVG paper)
scale_length <- 200
params <- data.frame(sigma.sq, beta)
plot(beta, sigma.sq)
#sampling from a poisson distribution - mean controls variance, so we don't specify tau.sq:
#step 1: use ST example distance matrix instead of creating a new one (Euclidean distance)
spe_demo <- Visium_humanDLPFC()
points_coord <- spatialCoords(spe_demo)
n_points <- nrow(points_coord)
pair.points <- cbind(
matrix( rep(points_coord, each = n_points), ncol = 2, byrow = FALSE),
rep(1, times = n_points) %x% points_coord # Creating the combinations using kronecker product.
) |> data.frame()
colnames(pair.points) <- c("si.x", "si.y", "sj.x", "sj.y")
#step 2: calculate gaussian process/kernel
kernel.fun <- function(si.x, si.y, sj.x, sj.y, l = 0.2){
exp(-1*sqrt(((si.x-sj.x)^2+(si.y-sj.y)^2))/l)
}
C_theta <- with(pair.points, kernel.fun(si.x, si.y, sj.x, sj.y, l = scale_length)) |>
matrix(nrow = n_points, ncol = n_points)
counts <- matrix(NA, nrow = n_genes, ncol = n_points)
eta_list <- list()
for (i in c(1:n_genes)) {
sigma.sq_i <- sigma.sq[i]
beta_i <- beta[i]
#step 3: simulate gaussian process per gene
gp_dat <- mvrnorm(n = 1, rep(0,n_points), sigma.sq_i* C_theta)
#step 4: calculate lambda = exp(beta + gaussian process) per gene
lambda_i <- exp(gp_dat + beta_i)
#step 5: use rpois() to simulate 4992 values per gene
counts_i <- rpois(n = n_points, lambda_i)
#put all counts in matrix
#orientation: genes x spots
counts[i,] <- counts_i
}
#create spe using counts and distance matrix
spe <- SpatialExperiment(
assays = list(counts = counts),
spatialCoords = points_coord)
rowData(spe)$ground_truth <- ground_truth
rowData(spe)$ground_truth_rank <- ground_truth_rank
rowData(spe)$ground_truth_sigma.sq <- sigma.sq
rowData(spe)$ground_truth_beta <- beta
Generate weights and SVG detection
library(SpatialExperiment)
library(nnSVG)
library(BRISC)
library(BiocParallel)
library(scuttle)
spe <- spe[, colSums(counts(spe)) > 0]
spe <- logNormCounts(spe)
weights <- generate_weights(input = spe, stabilize = TRUE)
spe_unweighted <- nnSVG(spe, assay_name = "logcounts")
spe_weighted <- weighted_nnSVG(input = spe, w = weights)
Visualize effect of weighting
library(ggplot2)
library(SpatialExperiment)
library(patchwork)
library(GGally)
library(dplyr)
library(ggridges)
#overlay unweighted and weighted ridge plots
df_unw <- data.frame(
rank = rowData(spe_unweighted)$rank,
mean = rowData(spe_unweighted)$mean,
method = rep("unw", 300)
) %>% mutate(quantile = findInterval(mean,
quantile(mean, probs=0:9/10))) %>%
tibble::rownames_to_column()
df_w <- data.frame(
rank = rowData(spe_weighted)$weighted_rank,
mean = rowData(spe_weighted)$weighted_mean,
method = rep("w", 300)
) %>% mutate(quantile = findInterval(mean,
quantile(mean, probs=0:9/10))) %>%
tibble::rownames_to_column()
df <- rbind(df_unw, df_w) %>%
mutate(quantile = as.factor(quantile))
ridge_overlay <- ggplot(df, aes(x = rank, y = quantile)) +
geom_density_ridges2(aes(fill = method), rel_min_height = 0.02, alpha = 0.3, scale = 1.3) +
theme_ridges(grid = TRUE) +
labs(
y = "decile - unw & w mean of logcounts",
x = "rank",
title = "Ridge plots: effect of weighting on rank"
) +
scale_fill_manual(labels = c("weighted", "unweighted"), values = c("blue", "red")) +
coord_cartesian(xlim = c(1, 300)) +
theme_bw()
#ridge plots separated by noise and signal for unweighted and weighted
frac <- round(dim(spe_unweighted)[1]*0.1)*0.1
df_unw_signal <- df_unw %>%
mutate(quantile = as.factor(quantile)) %>%
group_by(quantile) %>%
slice_min(order_by = rank, n = frac) %>%
mutate(grp = "signal")
indices <- as.integer(df_unw_signal$rowname)
df_unw_background <- df_unw[-indices,] %>%
mutate(quantile = as.factor(quantile)) %>%
mutate(grp = "background")
df <- rbind(df_unw_signal, df_unw_background)
rank_separated_unw <- ggplot(df, aes(x = rank, y = quantile)) +
geom_density_ridges2(aes(fill = grp), rel_min_height = 0.02, alpha = 0.3) +
theme_ridges(grid = TRUE) +
labs(
y = "decile - unw mean of logcounts",
x = "rank",
title = "Signal unweighted"
) +
guides(fill=guide_legend(title="group")) +
coord_cartesian(xlim = c(1, 300)) +
theme_bw()
df_w_signal <- df_w %>%
mutate(quantile = as.factor(quantile)) %>%
group_by(quantile) %>%
slice_min(order_by = rank, n = frac) %>%
mutate(grp = "signal")
indices <- as.integer(df_w_signal$rowname)
df_w_background <- df_w[-indices,] %>%
mutate(quantile = as.factor(quantile)) %>%
mutate(grp = "background")
df <- rbind(df_w_signal, df_w_background)
rank_separated_w <- ggplot(df, aes(x = rank, y = quantile)) +
geom_density_ridges2(aes(fill = grp), rel_min_height = 0.02, alpha = 0.3) +
theme_ridges(grid = TRUE) +
labs(
y = "decile - w mean of logcounts",
x = "rank",
title = "Signal weighted"
) +
guides(fill=guide_legend(title="group")) +
coord_cartesian(xlim = c(1, 300)) +
theme_bw()
ridge_signal <- wrap_plots(rank_separated_unw, rank_separated_w, nrow=1, guides = "collect")
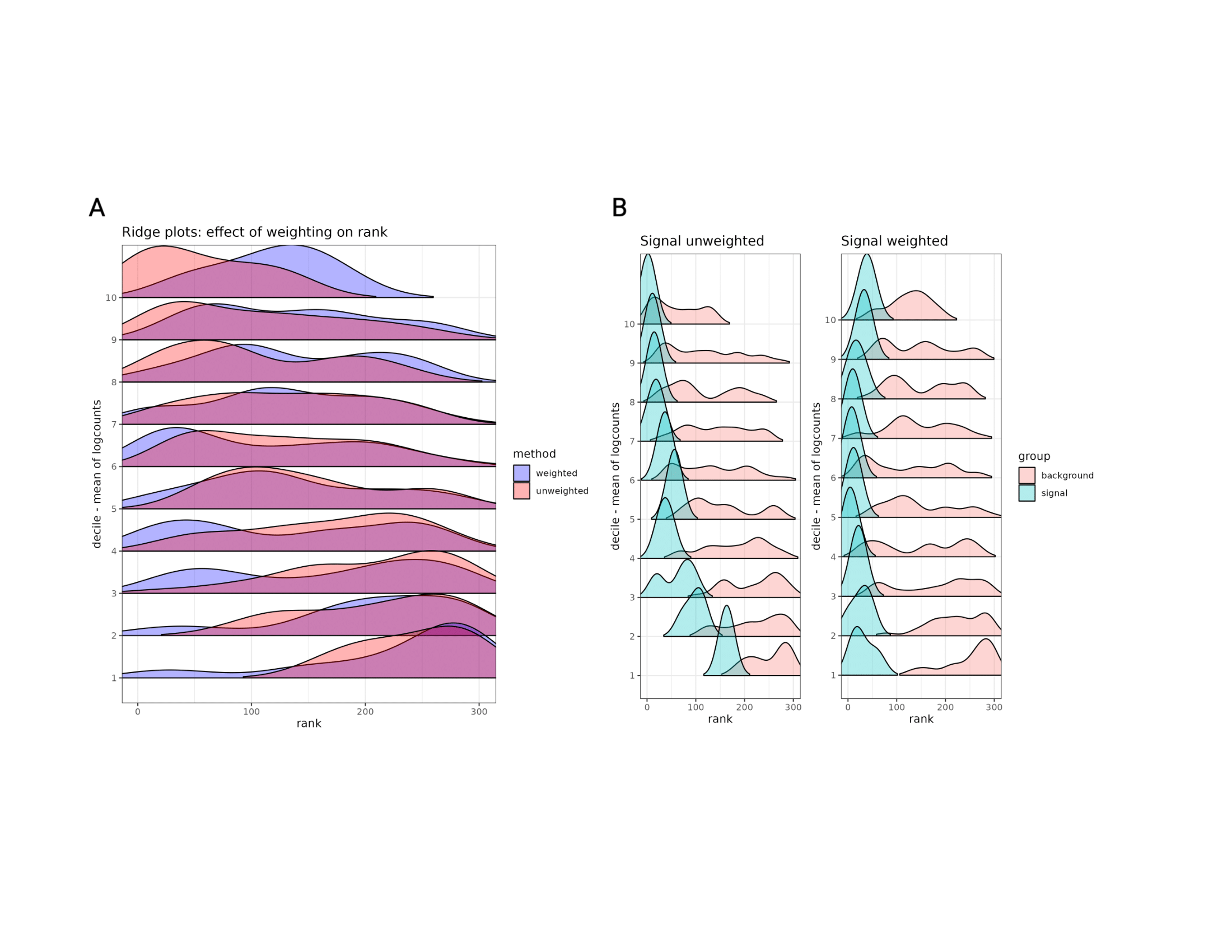
These code chunks have been evaluated and lead to the figure below with
both ridge plots. All the simulated genes are spatially varying genes,
but to different degrees, and the controlled mean and spatial variance
parameters are not correlated. After simulating the data, weights were
generated and constrained. To rank SVGs,
nnSVG was run on
both the unweighted logcounts and on the weighted logcounts matrices.
This figure shows the comparison between unweighted nnSVG ranks and
weighted nnSVG ranks within deciles based on mean expression. The
distribution of ranks moves toward zero for lower mean expression
deciles after the weighting is applied, indicating that SVGs are able to
be found even when they have low expression (subfigure A). Each decile
has spatially variable genes that should be highly ranked, and the
weighted method is able to recover these in the lowly expressed deciles.
In order to separate background noise from true signal, the densities of
the top three ranks from each decile are plotted separately from the
remaining genes in each decile (subfigure B). The weighted method is
able to find highly ranked SVGs even in lower deciles, showing that
spoon addresses the mean-variance relationship.
sessionInfo()