The goal of inlar is to ease output processing of INLA’s outputs by
putting them in tibble format.
You can install the released version of inlar here.
#' remotes::install_github("kklot/inlar")This is a basic example which shows you how to solve a common problem:
library(ggplot2)
library(inlar)
set.seed(1)The original data is binded to the output by default for those subject-specific outputs.
r = INLA::inla(y ~ 1 + f(x, model = 'iid'),
data = data.frame(y = rnorm(2), x = rnorm(2)),
control.compute = list(config = TRUE),
control.predictor = list(compute = TRUE, link = 1)
)
## Summary
r |> as_tibble(type = 'summary', 'random')
#> var id mean sd q025 q50 q975
#> 1 x -0.8356286 -0.4018769 0.3894734 -1.1826484 -0.4011349 0.3727972
#> 2 x 1.5952808 0.4018712 0.3894732 -0.3737118 0.4010382 1.1815156
r |> as_tibble(type = 'summary', 'linear.predictor')
#> # A tibble: 2 x 8
#> subj_id mean sd q025 q50 q975 y x
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Predictor.1 -0.624 0.0262 -0.661 -0.626 -0.573 -0.626 -0.836
#> 2 Predictor.2 0.181 0.0263 0.130 0.183 0.218 0.184 1.60
## Marginals
r |> as_tibble(type = 'marginals', 'random')
#> # A tibble: 150 x 4
#> var id x y
#> <chr> <dbl> <dbl> <dbl>
#> 1 x 1 -4.59 0.00000455
#> 2 x 1 -4.47 0.0000824
#> 3 x 1 -4.46 0.0000843
#> 4 x 1 -4.36 0.000102
#> 5 x 1 -3.80 0.000237
#> 6 x 1 -3.76 0.000254
#> 7 x 1 -3.00 0.000923
#> 8 x 1 -2.92 0.00107
#> 9 x 1 -2.62 0.00185
#> 10 x 1 -2.50 0.00236
#> # ... with 140 more rows
r |> as_tibble(type = 'marginals', 'linear.predictor')
#> # A tibble: 150 x 3
#> id x y
#> <dbl> <dbl> <dbl>
#> 1 1 -0.946 0.0000447
#> 2 1 -0.935 0.00107
#> 3 1 -0.935 0.00109
#> 4 1 -0.927 0.00129
#> 5 1 -0.881 0.00275
#> 6 1 -0.881 0.00276
#> 7 1 -0.817 0.00874
#> 8 1 -0.809 0.0103
#> 9 1 -0.785 0.0173
#> 10 1 -0.776 0.0212
#> # ... with 140 more rows
## Posterior sample
samples = inla.posterior.sample(2, r)
samples |> as_tibble("hyperpar")
#> # A tibble: 4 x 3
#> sample_id term value
#> <dbl> <chr> <dbl>
#> 1 1 Precision for the Gaussian observations 26680.
#> 2 1 Precision for x 4.74
#> 3 2 Precision for the Gaussian observations 6537.
#> 4 2 Precision for x 2.61
samples |> as_tibble("latent")
#> # A tibble: 10 x 4
#> sample_id term term_id value
#> <dbl> <chr> <dbl> <dbl>
#> 1 1 Predictor 1 -0.621
#> 2 1 Predictor 2 0.187
#> 3 1 x 1 -0.693
#> 4 1 x 2 0.113
#> 5 1 (Intercept) 1 0.0720
#> 6 2 Predictor 1 -0.606
#> 7 2 Predictor 2 0.160
#> 8 2 x 1 -1.05
#> 9 2 x 2 -0.283
#> 10 2 (Intercept) 1 0.441
samples |> as_tibble("logdens")
#> # A tibble: 2 x 4
#> sid hyperpar latent joint
#> <chr> <dbl> <dbl> <dbl>
#> 1 1 -14.1 17.5 3.41
#> 2 2 -13.1 12.2 -0.862Some figures
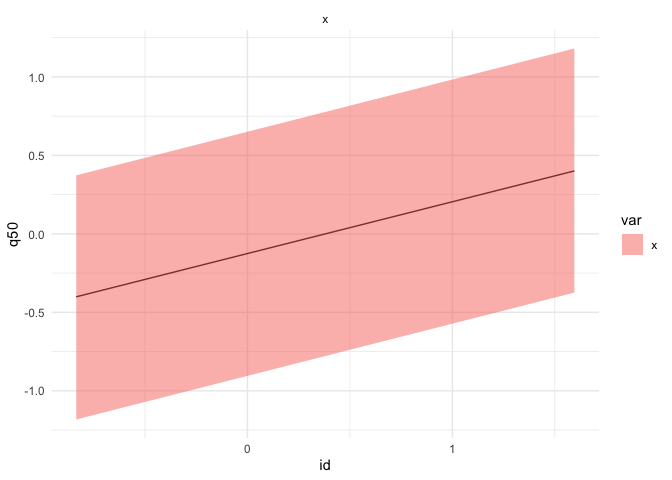
autoplot(r, "summary", "random") +
labs(title = "autoplot for INLA", x = "X's effect", y = "Est (95% CrI)")autoplot(r, "summary", "fitted.values") +
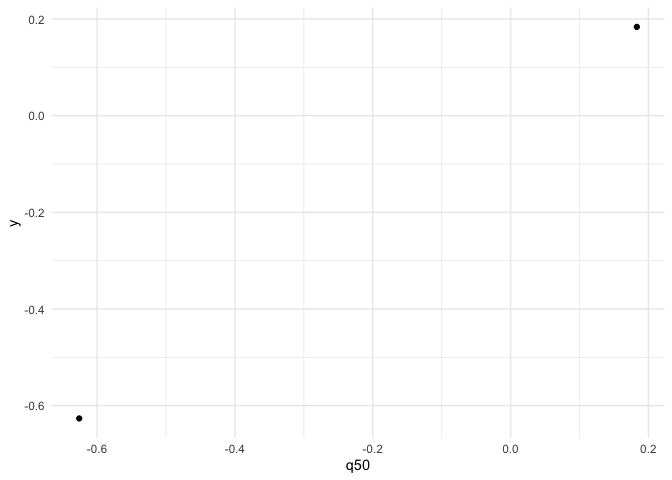
geom_abline(slope = 1, intercept = 0) +
labs(title = "Fitted vs data", x = "Fitted median", y = "Data", caption = 'can be wrong if link function is not correctly specify')