This package aims to make doing Bayesian things a little easier by giving a clear set of steps that you can easily use to simulate draws from a posterior distribution defined by a specific likelihood and prior.
definea likelihood
- Input: A data frame, a formula with the response variable and the name of a function that is your likelihood.
- Output: A data frame with the name of the function given at input as an attribute
assumea prior distribution
- Input: A data frame and a function of a single variable
- Ouput: A data frame with the name of the function added as an attribute
drawfrom the posterior distribution
- Input: A data frame, a control parameter
- Output: A data frame of draws from the posterior distribution
diagnosethe chain:
- Input: a data frame of draws from the posterior
- Output: The same data frame and a bunch of diagnostic plots
cleanthe chain
- Input: Draws from the posterior
- Output: Burned-in and subsampled draws from the posterior
- Construct your confidence interval or p-value
- Use
dplyrfor this
Estimate the standard deviation of a normal distribution with a gamma prior:
library(binfer)
library(tidyverse)
#> Loading tidyverse: ggplot2
#> Loading tidyverse: tibble
#> Loading tidyverse: tidyr
#> Loading tidyverse: readr
#> Loading tidyverse: purrr
#> Loading tidyverse: dplyr
#> Warning: package 'tibble' was built under R version 3.4.3
#> Conflicts with tidy packages ----------------------------------------------
#> filter(): dplyr, stats
#> lag(): dplyr, stats
my_lik <- function(data, theta) {if (theta > 0) dnorm(data, mean = mean(iris$Sepal.Width) , sd = theta) else 0}
my_prior <- function(theta) {dgamma(theta, shape = 10, rate = 20)}
posterior <- define(iris, Sepal.Width ~ my_lik) %>%
assume(prior = ~ my_prior) %>%
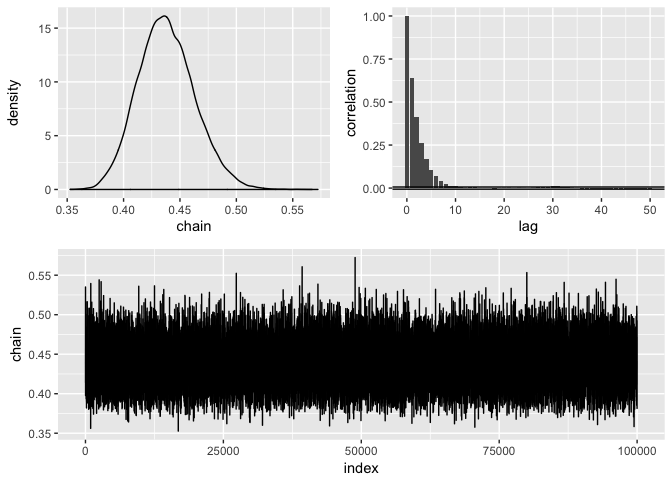
draw(initial = .43, nbatch = 1e5, blen = 1, scale = .05) %>%
diagnose() %>%
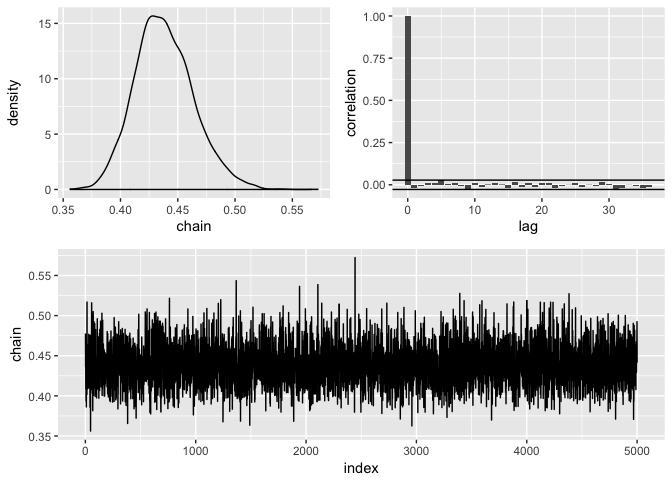
clean(burnin = 0, subsample = 20) %>%
diagnose()
#> Acceptance rate: 0.497534975349753
#> Acceptance rate: 1posterior %>% summarise(mean = mean(chain),
sd = sd(chain),
lower = quantile(chain, .025),
upper = quantile(chain, .975))
#> mean sd lower upper
#> 1 0.4382871 0.02558842 0.3925185 0.493049Estimate the probability of success of a binomnial distribution with a beta prior and compare the analytical solution to the approximate one:
binom_test_data <- rbinom(50, prob = .1, size = 1) %>%
tibble(response = .)
binom_lik <- function(data, theta) {if(theta > 0 & theta < 1) dbinom(data, prob = theta, size = 1) else 0}
beta_prior <- function(theta) {dbeta(theta, 1, 2)}
posterior2 <- binom_test_data %>%
define(response ~ binom_lik) %>%
assume(~ beta_prior) %>%
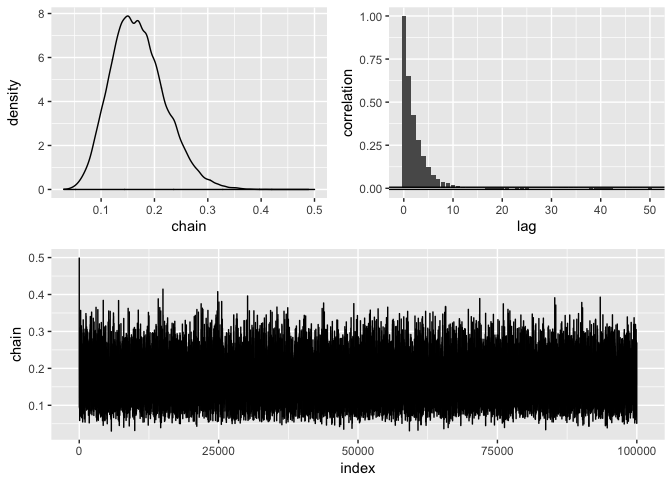
draw(initial = .5, nbatch = 1e5, blen = 1, scale = .15) %>%
diagnose() %>%
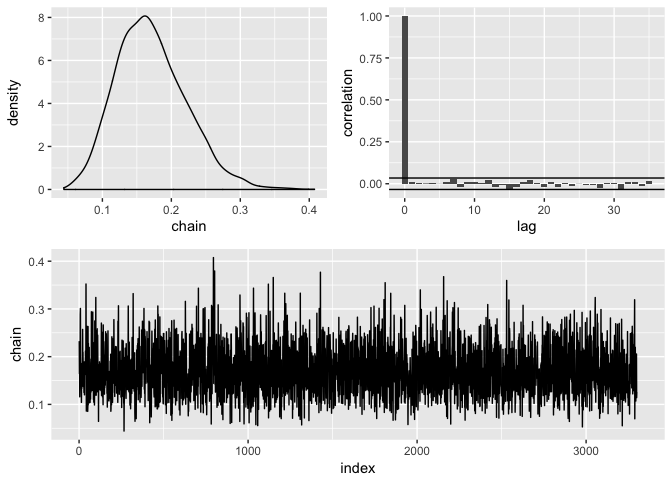
clean(burnin = 1000, subsample = 30) %>%
diagnose()
#> Acceptance rate: 0.376063760637606
#> Acceptance rate: 1posterior2 %>%
summarize(mean = mean(chain),
sd = sd(chain),
lower = quantile(chain, .025),
upper = quantile(chain, .975))
#> # A tibble: 1 x 4
#> mean sd lower upper
#> <dbl> <dbl> <dbl> <dbl>
#> 1 0.171 0.0508 0.0853 0.282
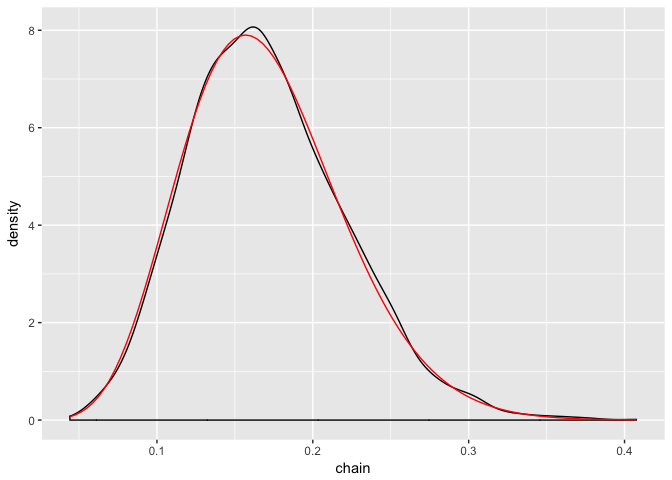
ggplot(posterior2, aes(chain)) +
geom_density() +
stat_function(fun = dbeta,
args = list(1 + sum(binom_test_data$response),
2 + nrow(binom_test_data) - sum(binom_test_data$response)),
color = "red")