Lightweight data management and analysis tools for single-molecule microscopy.
Table of Contents generated with DocToc
The B-Store documentation is found at Read the Docs: http://b-store.readthedocs.org.
Examples for how to use B-Store are located in the examples folder.
If you need help, post a question to the B-Store Google Groups discussion page: https://groups.google.com/forum/#!forum/b-store
Bug reports may be submitted to the GitHub issue tracker: https://github.com/kmdouglass/bstore/issues
If you find B-Store useful, please consider citing it by referencing the DOI of the specific version in use. For example:
Douglass, Kyle M., Sieben, Christian, Berliner, Niklas, & Manley, Suliana. (2017, December 18). B-Store (Version v1.2.1). Zenodo. https://doi.org/10.5281/zenodo.1117843
B-Store is most easily installed from the Anaconda Cloud package repository. If you don't already have Anaconda installed, you may download it for Python 3.5 and greater from https://www.continuum.io/downloads. Once installed, run the commands from the terminal listed below for your system. (If you're on Windows, use the Anaconda Prompt that is supplied with Anaconda.)
Note that these commands will install B-Store into an environment named bstore that is independent of your default environment. When you want to activate this environment to use B-Store, simply type source activate bstore in the Linux/OSX terminal or activate bstore in the Windows Anaconda Prompt.
conda update conda
conda config --append channels soft-matter
conda create -n bstore -c kmdouglass bstoreTo install from source, simply clone this repository and install B-Store using pip.
git clone https://github.com/kmdouglass/bstore.git
pip install bstoreThe most up-to-date code may be found on the development branch, though it may not be as thoroughly tested as code on the master. Source installs will always be more up-to-date than the Anaconda packages.
A partial list of requirements to run B-Store may be found in the requirements.txt file.
If you installed B-Store using the Anaconda package manager, updating it is easy. The following assumes that the soft-matter and conda-forge channels were appended to your conda config as described above. It also assumes that you have already activated the environment in which you installed B-Store.
conda update -c kmdouglass bstoreIf instead you installed B-Store from source using pip, simply uninstall your current version, download or pull the latest code from the GitHub repository, then install the new version as before.
The Anaconda package manager can sometimes have difficulties when
finding the most up-to-date version of B-Store for Windows x64 (the
same problem has not been observed on Linux x64 systems). This will be
apparent when, using the directions listed above, conda will ask you
to verify the version of the packages to install. If during the
install conda indicates that it wants to install a version of
B-Store that is less than the most up-to-date version listed at the
top of this README file, then try downgrading conda. Version 4.2.x has
been known to fix this problem, whereas versions 4.3.7 and 4.3.8 have
been found to cause the problem.
In the root conda environment, enter the command:
conda install conda=4.2.16B-Store is a lightweight data management and analysis library for single molecule localization microscopy (SMLM). It serves two primary roles:
- To structure SMLM data inside a database for fast and easy information retrieval and storage.
- To facilitate the analysis of high-throughput SMLM datasets.
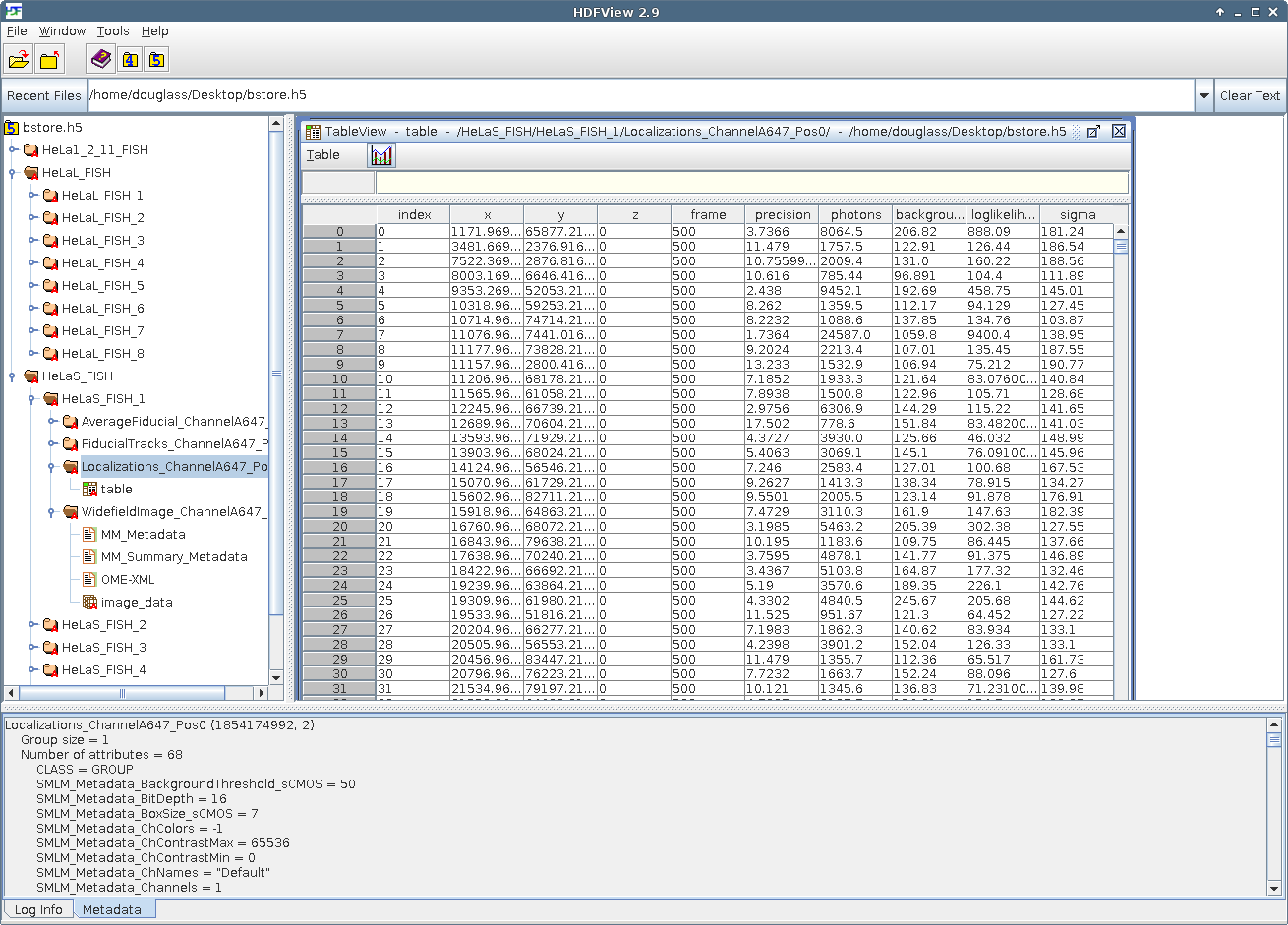
As an example, you can see how SMLM data is organized by B-Store inside a database in the HDF format below. (The software used to view the database is not B-Store, but HDFView from the HDF Group.)
Please see the FAQ for more information.