Latent Dirichlet Allocation Using ‘tidyverse’ Conventions
Copyright 2020 by Thomas W. Jones
Implements an algorithm for Latent Dirichlet Allocation using style conventions from the tidyverse and tidymodels.
In addition this implementation of LDA allows you to:
- use asymmetric prior parameters alpha and beta
- use a matrix prior parameter, beta to seed topics into a model
- use a previously-trained model as a prior for a new model
- apply LDA in a transfer-learning paradigm, updating a model’s parameters with additional data (or additional iterations)
Note that the seeding of topics and transfer learning are experimental for now. They are almost-surely useful but their behaviors have not been optimized or well-studied. Caveat emptor!
You can install the development version from GitHub with:
install.packages("remotes")
remotes::install_github("tommyjones/tidylda")This package is still in its early stages of development. However, some
basic functionality is below. Here, we will use the tidytext package
to create a document term matrix, fit a topic model, predict topics of
unseen documents, and update the model with those new documents.
tidylda uses the following naming conventions for topic models:
thetais a matrix whose rows are distributions of topics over documents, or P(topic|document)phiis a matrix whose rows are distributions of tokens over topics, or P(token|topic)gammais a matrix whose rows are distributions of topics over tokens, or P(topic|token)gammais useful for making predictions with a computationally-simple and efficient dot product and it may be interesting to analyze in its own right.alphais the prior that tunesthetabetais the prior that tunesphi
library(tidytext)
library(tidyverse)
#> ── Attaching packages ──────────────────────────────────────────────────────────────── tidyverse 1.3.0 ──
#> ✓ ggplot2 3.3.0 ✓ purrr 0.3.4
#> ✓ tibble 3.0.1 ✓ dplyr 1.0.0
#> ✓ tidyr 1.1.0 ✓ stringr 1.4.0
#> ✓ readr 1.3.1 ✓ forcats 0.5.0
#> ── Conflicts ─────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
#> x dplyr::filter() masks stats::filter()
#> x dplyr::lag() masks stats::lag()
library(tidylda)
#> Registered S3 method overwritten by 'tidylda':
#> method from
#> tidy.matrix broom
library(Matrix)
#>
#> Attaching package: 'Matrix'
#> The following objects are masked from 'package:tidyr':
#>
#> expand, pack, unpack
### Initial set up ---
# load some documents
docs <- textmineR::nih_sample
# tokenize using tidytext's unnest_tokens
tidy_docs <- docs %>%
select(APPLICATION_ID, ABSTRACT_TEXT) %>%
unnest_tokens(output = word,
input = ABSTRACT_TEXT,
stopwords = stop_words$word,
token = "ngrams",
n_min = 1, n = 2) %>%
count(APPLICATION_ID, word) %>%
filter(n>1) #Filtering for words/bigrams per document, rather than per corpus
tidy_docs <- tidy_docs %>% # filter words that are just numbers
filter(! stringr::str_detect(tidy_docs$word, "^[0-9]+$"))
# turn a tidy tbl into a sparse dgCMatrix
# note tidylda has support for several document term matrix formats
d <- tidy_docs %>%
cast_sparse(APPLICATION_ID, word, n)
# let's split the documents into two groups to demonstrate predictions and updates
d1 <- d[1:50, ]
d2 <- d[51:nrow(d), ]
# make sure we have different vocabulary for each data set to simulate the "real world"
# where you get new tokens coming in over time
d1 <- d1[, colSums(d1) > 0]
d2 <- d2[, colSums(d2) > 0]
### fit an intial model and inspect it ----
set.seed(123)
lda <- tidylda(
dtm = d1,
k = 10,
iterations = 200,
burnin = 175,
alpha = 0.1, # also accepts vector inputs
beta = 0.05, # also accepts vector or matrix inputs
optimize_alpha = FALSE, # experimental
calc_likelihood = TRUE,
calc_r2 = TRUE, # see https://arxiv.org/abs/1911.11061
return_data = FALSE
)
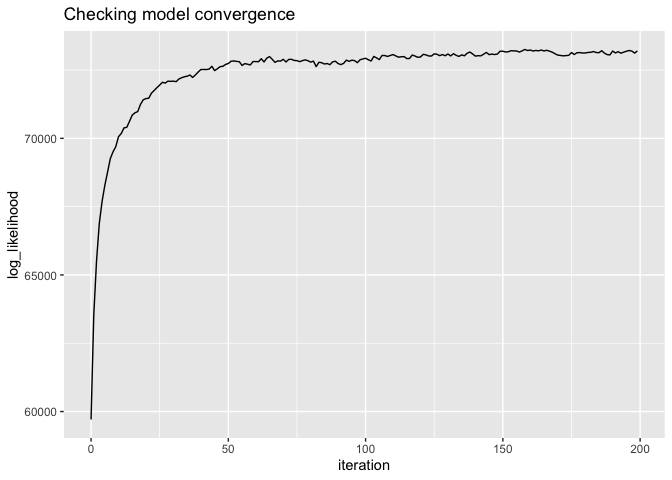
# did the model converge?
# there are actual test stats for this, but should look like "yes"
qplot(x = iteration, y = log_likelihood, data = lda$log_likelihood, geom = "line") +
ggtitle("Checking model convergence")# look at the model overall
glance(lda)
#> # A tibble: 1 x 5
#> num_topics num_documents num_tokens iterations burnin
#> <int> <int> <int> <dbl> <dbl>
#> 1 10 50 1597 200 175
print(lda)
#> A Latent Dirichlet Allocation Model of 10 topics, 50 documents, and 1597 tokens:
#> tidylda(dtm = d1, k = 10, iterations = 200, burnin = 175, alpha = 0.1,
#> beta = 0.05, optimize_alpha = FALSE, calc_likelihood = TRUE,
#> calc_r2 = TRUE, return_data = FALSE)
#>
#> The model's R-squared is 0.2812
#> The 5 most prevalent topics are:
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 6 12 0.290 imaging, diabetes, cancer, ...
#> 2 1 11.4 0.218 cell, specific, in, ...
#> 3 9 11.4 0.272 v4, effects, stiffening, ...
#> 4 2 11.3 0.118 mitochondrial, the, cells, ...
#> 5 4 9.95 0.426 c, function, cmybp, ...
#> # … with 5 more rows
#>
#> The 5 most coherent topics are:
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 4 9.95 0.426 c, function, cmybp, ...
#> 2 7 7.73 0.303 sud, risk, cdk5, ...
#> 3 6 12 0.290 imaging, diabetes, cancer, ...
#> 4 9 11.4 0.272 v4, effects, stiffening, ...
#> 5 1 11.4 0.218 cell, specific, in, ...
#> # … with 5 more rows
# it comes with its own summary matrix that's printed out with print(), above
lda$summary
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 1 11.4 0.218 cell, specific, in, ...
#> 2 2 11.3 0.118 mitochondrial, the, cells, ...
#> 3 3 9.61 0.0257 sleep, the, repair, ...
#> 4 4 9.95 0.426 c, function, cmybp, ...
#> 5 5 7.83 0.0840 the, cns, pd, ...
#> 6 6 12 0.290 imaging, diabetes, cancer, ...
#> 7 7 7.73 0.303 sud, risk, cdk5, ...
#> 8 8 9.91 0.0725 program, dcis, the, ...
#> 9 9 11.4 0.272 v4, effects, stiffening, ...
#> 10 10 8.91 0.0846 the, research, core, ...
# inspect the individual matrices
tidy_theta <- tidy(lda, matrix = "theta")
tidy_theta
#> # A tibble: 500 x 3
#> document topic theta
#> <chr> <dbl> <dbl>
#> 1 8574224 1 0.00739
#> 2 8574224 2 0.00217
#> 3 8574224 3 0.00304
#> 4 8574224 4 0.00217
#> 5 8574224 5 0.00304
#> 6 8574224 6 0.00217
#> 7 8574224 7 0.149
#> 8 8574224 8 0.00478
#> 9 8574224 9 0.0196
#> 10 8574224 10 0.807
#> # … with 490 more rows
tidy_phi <- tidy(lda, matrix = "phi")
tidy_phi
#> # A tibble: 15,970 x 3
#> topic token phi
#> <dbl> <chr> <dbl>
#> 1 1 adolescence 0.0000599
#> 2 1 age 0.0000599
#> 3 1 application 0.0000599
#> 4 1 depressive 0.0000599
#> 5 1 disorder 0.0000599
#> 6 1 emotionality 0.0000599
#> 7 1 information 0.0000599
#> 8 1 mdd 0.0000599
#> 9 1 onset 0.0000599
#> 10 1 onset.mdd 0.0000599
#> # … with 15,960 more rows
tidy_gamma <- tidy(lda, matrix = "gamma")
tidy_gamma
#> # A tibble: 15,970 x 3
#> topic token gamma
#> <dbl> <chr> <dbl>
#> 1 1 adolescence 0.00794
#> 2 1 age 0.00935
#> 3 1 application 0.00785
#> 4 1 depressive 0.0205
#> 5 1 disorder 0.0205
#> 6 1 emotionality 0.0205
#> 7 1 information 0.00671
#> 8 1 mdd 0.0114
#> 9 1 onset 0.00800
#> 10 1 onset.mdd 0.0206
#> # … with 15,960 more rows
### predictions on held out data ---
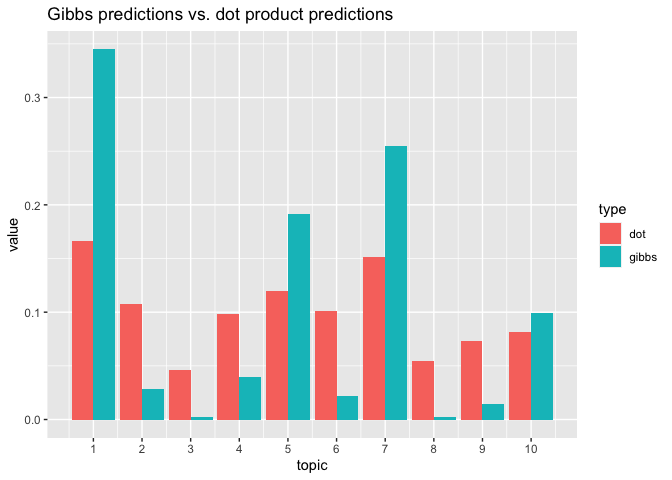
# two methods: gibbs is cleaner and more techically correct in the bayesian sense
p_gibbs <- predict(lda, new_data = d2[1, ], iterations = 100, burnin = 75)
# dot is faster, less prone to error (e.g. underflow), noisier, and frequentist
p_dot <- predict(lda, new_data = d2[1, ], method = "dot")
# pull both together into a plot to compare
tibble(topic = 1:ncol(p_gibbs), gibbs = p_gibbs[1,], dot = p_dot[1, ]) %>%
pivot_longer(cols = gibbs:dot, names_to = "type") %>%
ggplot() +
geom_bar(mapping = aes(x = topic, y = value, group = type, fill = type),
stat = "identity", position="dodge") +
scale_x_continuous(breaks = 1:10, labels = 1:10) +
ggtitle("Gibbs predictions vs. dot product predictions")### updating the model ----
# now that you have new documents, maybe you want to fold them into the model?
lda2 <- refit(
object = lda,
dtm = d, # save me the trouble of manually-combining these by just using d
iterations = 200,
burnin = 175,
calc_likelihood = TRUE,
calc_r2 = TRUE
)
# we can do similar analyses
# did the model converge?
qplot(x = iteration, y = log_likelihood, data = lda2$log_likelihood, geom = "line") +
ggtitle("Checking model convergence")# look at the model overall
glance(lda2)
#> # A tibble: 1 x 5
#> num_topics num_documents num_tokens iterations burnin
#> <int> <int> <int> <dbl> <dbl>
#> 1 10 99 3081 200 175
print(lda2)
#> A Latent Dirichlet Allocation Model of 10 topics, 99 documents, and 3081 tokens:
#> refit.tidylda(object = lda, dtm = d, iterations = 200, burnin = 175,
#> calc_likelihood = TRUE, calc_r2 = TRUE)
#>
#> The model's R-squared is 0.1644
#> The 5 most prevalent topics are:
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 8 12.6 0.107 research, the, program, ...
#> 2 1 11.5 0.192 cell, cells, infection, ...
#> 3 2 11.0 0.140 hiv, the, mitochondrial, ...
#> 4 3 10.8 0.0212 microbiome, ptc, gut, ...
#> 5 5 9.57 0.0209 the, treatment, disease, ...
#> # … with 5 more rows
#>
#> The 5 most coherent topics are:
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 7 9.06 0.256 risk, factors, sud, ...
#> 2 4 8.04 0.198 c, function, determine, ...
#> 3 1 11.5 0.192 cell, cells, infection, ...
#> 4 2 11.0 0.140 hiv, the, mitochondrial, ...
#> 5 6 8.97 0.112 cancer, cells, imaging, ...
#> # … with 5 more rows
# how does that compare to the old model?
print(lda)
#> A Latent Dirichlet Allocation Model of 10 topics, 50 documents, and 1597 tokens:
#> tidylda(dtm = d1, k = 10, iterations = 200, burnin = 175, alpha = 0.1,
#> beta = 0.05, optimize_alpha = FALSE, calc_likelihood = TRUE,
#> calc_r2 = TRUE, return_data = FALSE)
#>
#> The model's R-squared is 0.2812
#> The 5 most prevalent topics are:
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 6 12 0.290 imaging, diabetes, cancer, ...
#> 2 1 11.4 0.218 cell, specific, in, ...
#> 3 9 11.4 0.272 v4, effects, stiffening, ...
#> 4 2 11.3 0.118 mitochondrial, the, cells, ...
#> 5 4 9.95 0.426 c, function, cmybp, ...
#> # … with 5 more rows
#>
#> The 5 most coherent topics are:
#> # A tibble: 10 x 4
#> topic prevalence coherence top_terms
#> <dbl> <dbl> <dbl> <chr>
#> 1 4 9.95 0.426 c, function, cmybp, ...
#> 2 7 7.73 0.303 sud, risk, cdk5, ...
#> 3 6 12 0.290 imaging, diabetes, cancer, ...
#> 4 9 11.4 0.272 v4, effects, stiffening, ...
#> 5 1 11.4 0.218 cell, specific, in, ...
#> # … with 5 more rowsI plan to have more analyses and a fuller accounting of the options of the various functions when I write the vignettes.
Planned updates include:
- an
augmentmethod to append distributions oftheta,phi, orgammato a tidy tibble of tokens - various functions to compare topic models to evaluate the effects of
refit. (Although the functions will likely be general enough that you could compare any topic models.)
If you have any suggestions for additional functionality, changes to functionality, changes to arguments or other aspects of the API please let me know by opening an issue or sending me an email: jones.thos.w at gmail.com.