ggrapid: Create neat & complete ggplot visualizations with as little code as possible
Overview
ggrapid enables creation of the most common ggplot-based visualizations fast and with just a few lines of code. In practice the package offers wrappers of some of the most common ggplot geoms such as: geom_density, geom_boxplot, geom_bar etc. ggrapid comes handy when you’d like to do an initial and quick EDA (Exploratory Data Analysis) over various columns of your dataset programatically, without the need of writing a lot of custom ggplot code.
Installation
# Install development version from GitHub
devtools::install_github("konradsemsch/ggrapid")Main functions
ggrapid offers a couple wrappers around the most commonly used functions in the course of doing an EDA:
plot_densityplot_boxplotplot_deciles(withcalculate_decile_table)plot_correlationplot_barsplot_line
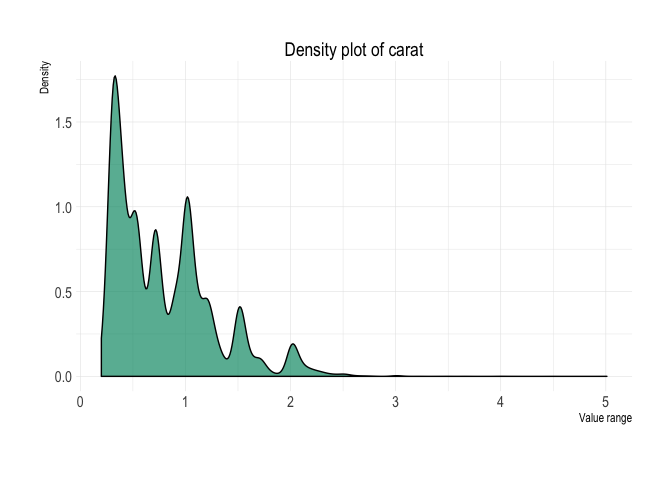
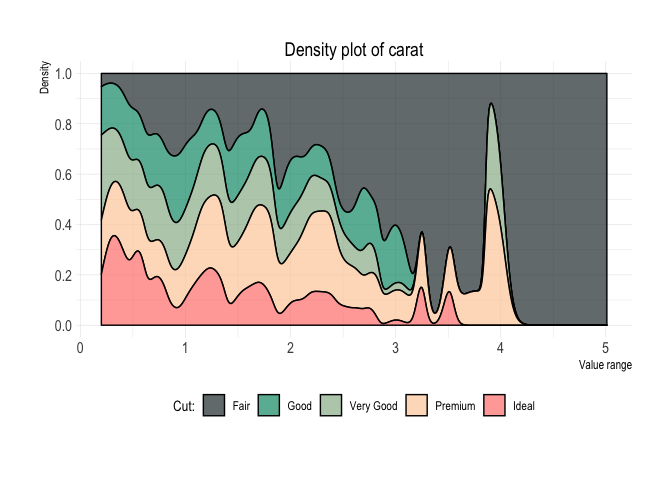
Density plot
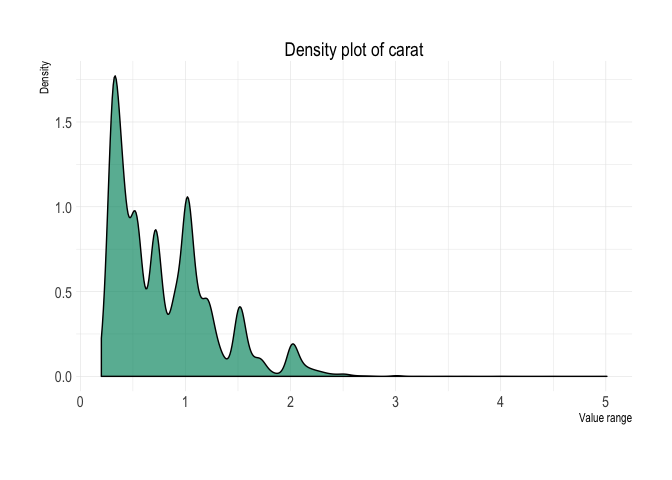
diamonds %>%
plot_density(x = carat)Box-plot
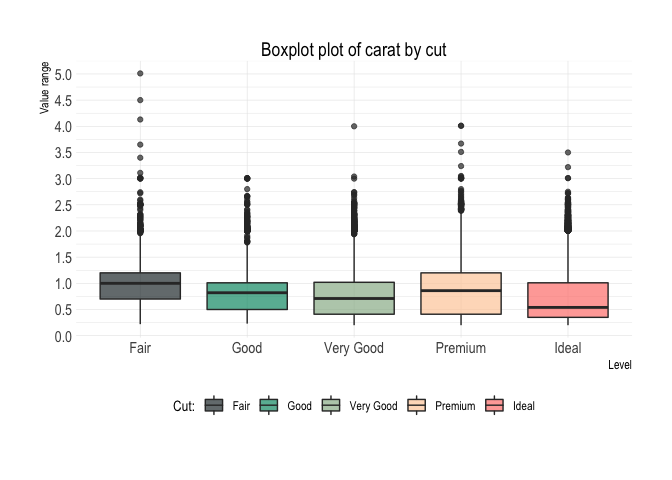
diamonds %>%
plot_boxplot(x = cut,
y = carat,
fill = cut)Decile plot
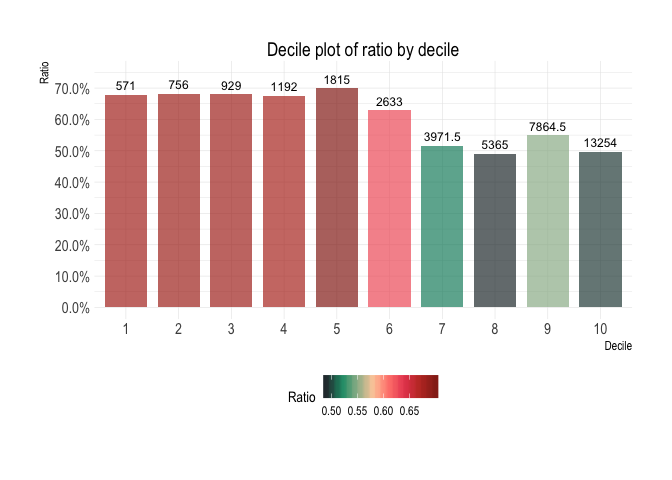
diamonds %>%
filter(cut %in% c("Ideal", "Premium")) %>%
calculate_decile_table(price, cut, "Ideal") %>%
plot_deciles()Correlation
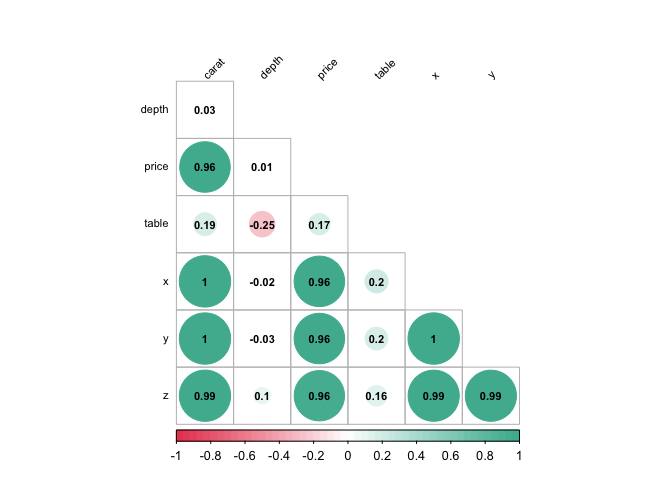
diamonds %>%
plot_correlation()Barplot
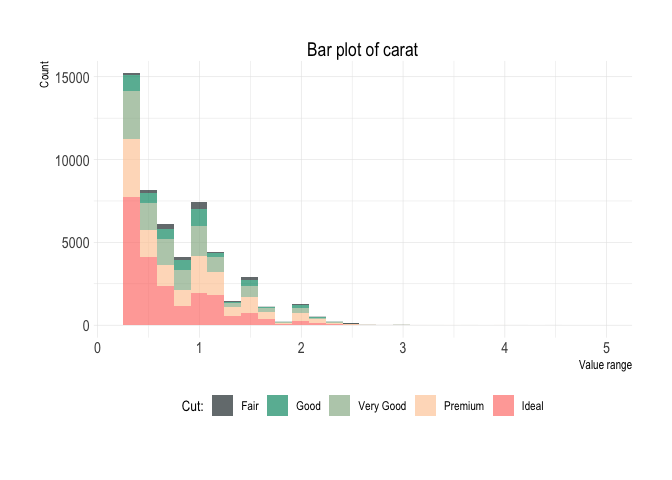
diamonds %>%
plot_bars(x = carat,
x_type = "num",
fill = cut)Lineplot
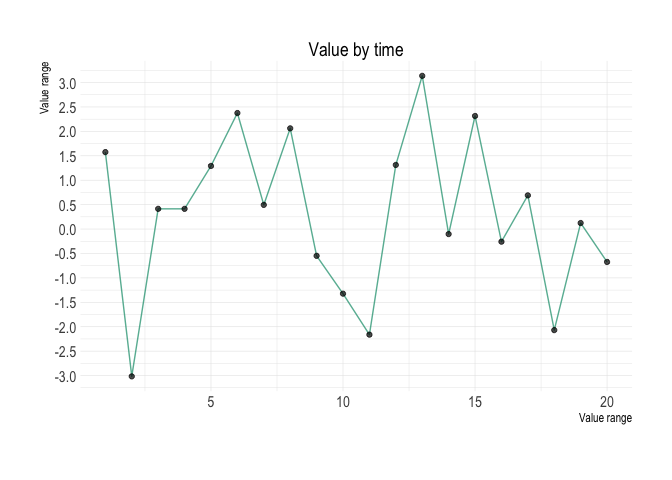
tibble(
time = 1:20,
value = rnorm(20, 0.5, 2)
) %>%
plot_line(
x = time,
y = value
)Main arguments
The most commonly implemented ggplot2 arguments across all main ggrapid functions ensure that you can build your basic EDA file without making additional changes or custom functions. Those arguments are mainly (might slightly differ across functions):
- fill
- facet
- position
- ticks
- angle
- title
- subtitle
- caption
- lab_x
- lab_y
- legend
- vline/ hline
- alpha
- quantile_low
- quantile_high
- theme_type
- palette
diamonds %>%
plot_density(x = carat)diamonds %>%
plot_density(x = carat,
fill = cut,
position = "stack")diamonds %>%
plot_density(x = carat,
fill = cut,
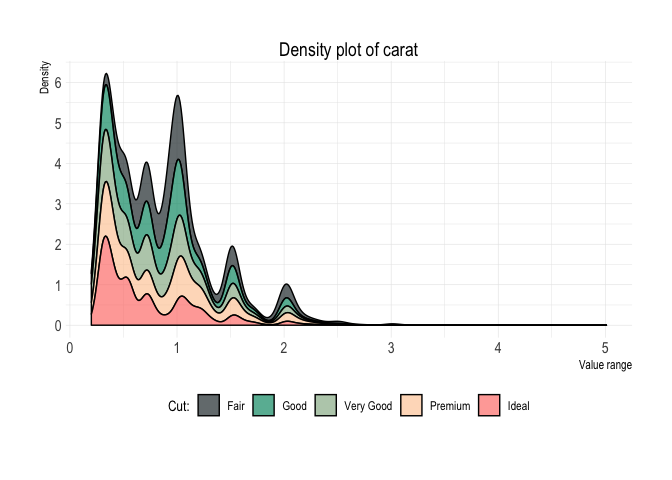
position = "fill")diamonds %>%
plot_density(x = carat,
fill = cut,
facet = cut,
title = "Write your title here",
subtitle = "Write your subtitle here",
caption = "Write your caption here",
lab_x = "Carat",
alpha = .5,
vline = 1)Complete usage
You can easily iterate across selected columns and create a set of plots for your EDA file:
library(recipes)
credit_data_nested <- credit_data %>%
select(-one_of("Home", "Marital", "Records", "Job")) %>% # removing categorical variables
gather(variable, variable_value,
one_of("Seniority", "Time", "Age", "Expenses", # selecting variables to gather
"Income", "Assets", "Debt", "Amount", "Price")) %>%
nest(-variable) %>%
mutate(
decile_table = map(data,
~calculate_decile_table(
.x,
binning = variable_value,
grouping = Status,
top_level = "bad",
format = FALSE
)
),
plot_deciles = pmap(list(x = decile_table, y = variable),
~plot_deciles(
.x,
title = glue::glue("Decile plot of {.y}"),
quantile_low = 0,
quantile_high = 1,
lab_x = "Decile",
lab_y = "Bad rate, %"
)
),
plot_boxplot = pmap(list(x = data, y = variable),
~plot_boxplot(
.x,
x = Status,
y = variable_value,
fill = Status,
title = glue::glue("Box plot of {.y} by Status"),
quantile_low = 0.01,
quantile_high = 0.99,
lab_x = "Performance",
caption = "Removed 1% of observations from each side",
palette = "inv_binary"
)
),
plot_density = pmap(list(x = data, y = variable),
~plot_density(
.x,
x = variable_value,
fill = Status,
title = glue::glue("Box plot of {.y} by Status"),
quantile_low = 0.01,
quantile_high = 0.99,
lab_x = "Performance",
caption = "Removed 1% of observations from each side",
palette = "inv_binary"
)
)
)This will give you the following structure. Each row represents an individual variable and columns are the different plots you would like to inspect:
credit_data_nested[1:3, ]
#> # A tibble: 3 x 6
#> variable data decile_table plot_deciles plot_boxplot plot_density
#> <chr> <list> <list> <list> <list> <list>
#> 1 Seniority <tibble [… <tibble [10 … <gg> <gg> <gg>
#> 2 Time <tibble [… <tibble [10 … <gg> <gg> <gg>
#> 3 Age <tibble [… <tibble [10 … <gg> <gg> <gg>Exemplary EDA format
Creating a standardised EDA file is just as easy as doing something like this:
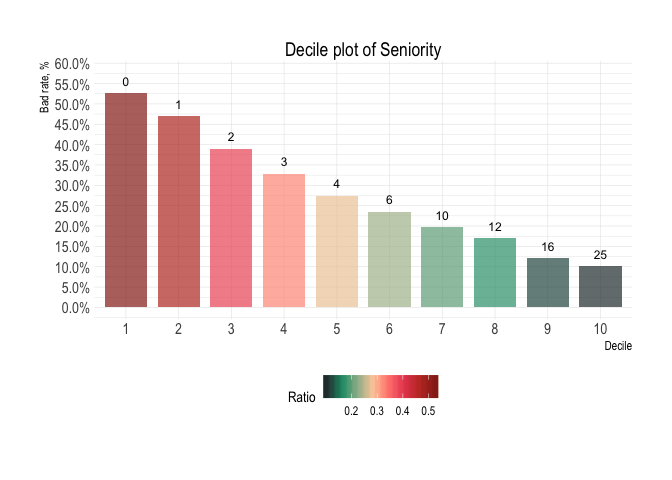
Variable: Seniority
Decile analysis
credit_data_nested$decile_table[[1]]
#> # A tibble: 10 x 8
#> decile min median max top_level total bottom_level ratio
#> <fct> <dbl> <dbl> <dbl> <int> <int> <int> <formttbl>
#> 1 1 0 0 0 235 446 211 52.69%
#> 2 2 0 1 1 209 445 236 46.97%
#> 3 3 1 2 2 174 446 272 39.01%
#> 4 4 2 3 3 146 445 299 32.81%
#> 5 5 3 4 5 122 445 323 27.42%
#> 6 6 5 6 8 105 446 341 23.54%
#> 7 7 8 10 10 88 445 357 19.78%
#> 8 8 10 12 14 76 446 370 17.04%
#> 9 9 14 16 20 54 445 391 12.13%
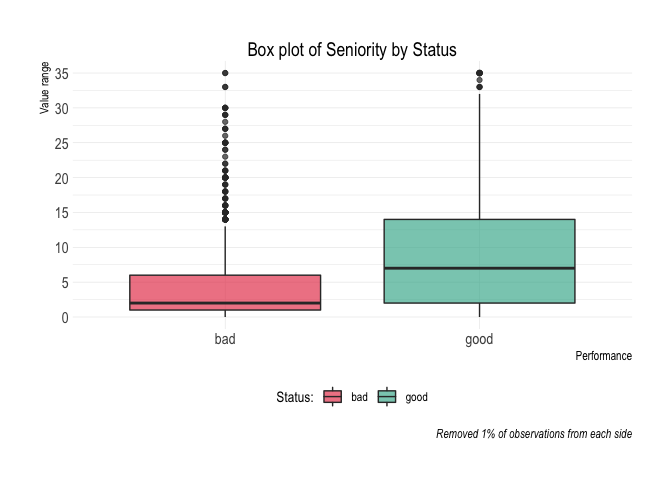
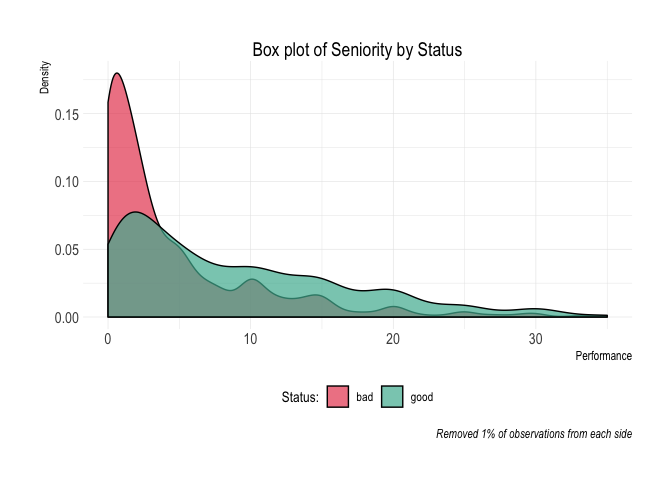
#> 10 10 20 25 48 45 445 400 10.11%credit_data_nested$plot_deciles[[1]]Aditional plots
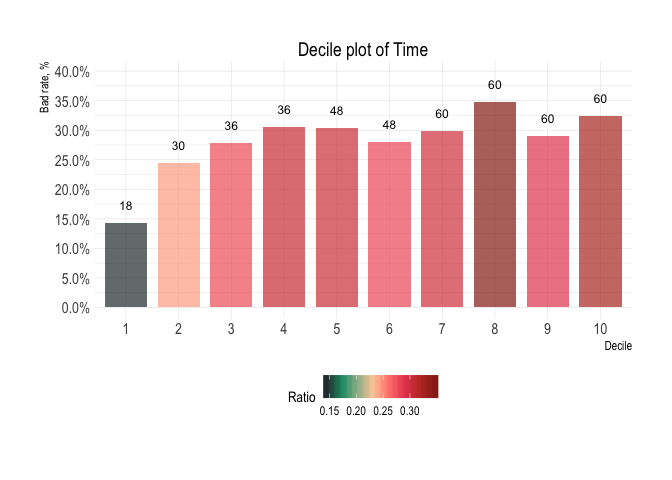
credit_data_nested$plot_boxplot[[1]]credit_data_nested$plot_density[[1]]Variable: Time
Decile analysis
credit_data_nested$decile_table[[2]]
#> # A tibble: 10 x 8
#> decile min median max top_level total bottom_level ratio
#> <fct> <dbl> <dbl> <dbl> <int> <int> <int> <formttbl>
#> 1 1 6 18 24 64 446 382 14.35%
#> 2 2 24 30 36 109 445 336 24.49%
#> 3 3 36 36 36 124 446 322 27.80%
#> 4 4 36 36 48 136 445 309 30.56%
#> 5 5 48 48 48 135 445 310 30.34%
#> 6 6 48 48 60 125 446 321 28.03%
#> 7 7 60 60 60 133 445 312 29.89%
#> 8 8 60 60 60 155 446 291 34.75%
#> 9 9 60 60 60 129 445 316 28.99%
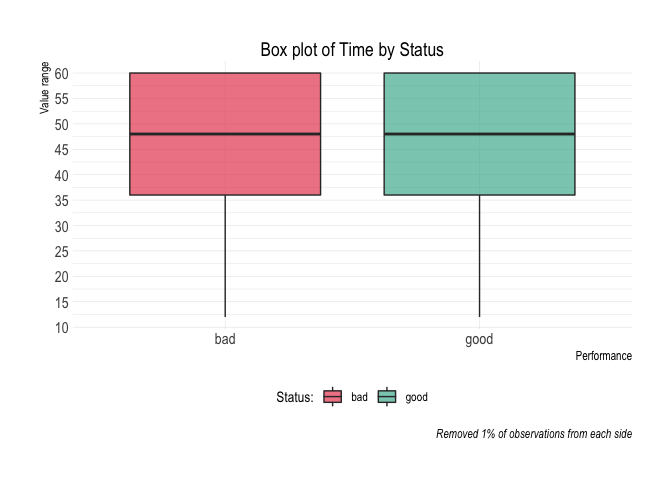
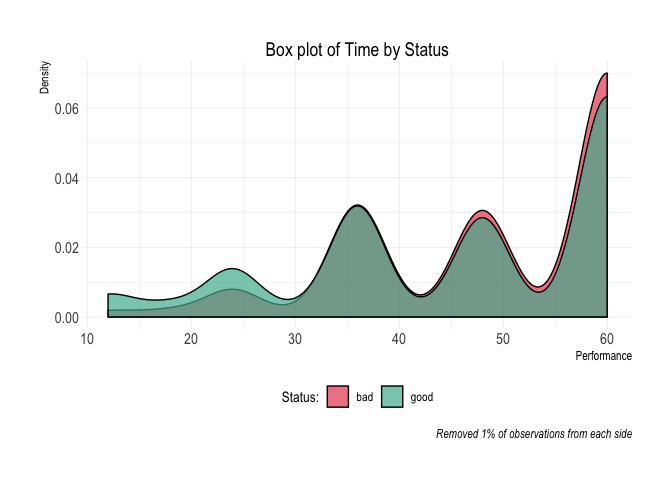
#> 10 10 60 60 72 144 445 301 32.36%credit_data_nested$plot_deciles[[2]]Aditional plots
credit_data_nested$plot_boxplot[[2]]credit_data_nested$plot_density[[2]]