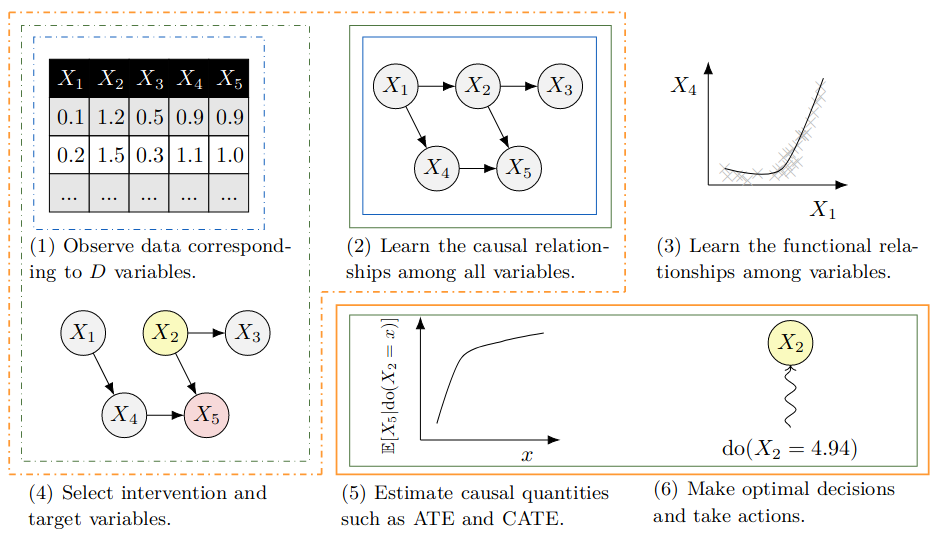
The task of Causal Discovery is to uncover the true DAG
In this project, we select DECI (Deep End-to-end Causal Inference), a SOTA causal discovery algorithm for iid observational data by [GAF+22], a variational inference model modeling exogenous noise as a normalizing flow.
Since the literature on causal discovery algorithms for time-series data using deep learning techniques is quite limited and virginal, we opt to implement DECI on time-series data using lagged cross-sectional data. Finally, we evaluate our performance against non-deep-learning-inspired algorithms (PCMCI, PCMCI(+) etc.) on time-series synthetic data with known ground truth causal graph [LKSS20].
- [GAF+22] Tomas Geffner, Javier Antoran, Adam Foster, Wenbo Gong, Chao Ma, Emre Kiciman, Amit Sharma, Angus Lamb, Martin Kukla, Nick Pawlowski, et al. Deep end-to-end causal inference. arXiv preprint arXiv:2202.02195, 2022.
- [LKSS20] Andrew R. Lawrence, Marcus Kaiser, Rui Sampaio, and Maksim Sipos. Data generating process to evaluate causal discovery techniques for time series data. Causal Discovery Causality-Inspired Machine Learning Workshop at Neural Information Processing Systems, 2020.
- [VCB22] Matthew J Vowels, Necati Cihan Camgoz, and Richard Bowden. D’ya like DAGs? a survey on structure learning and causal discovery. ACM Computing Surveys, 55(4):1–36, 2022.
- [ZARX18] Xun Zheng, Bryon Aragam, Pradeep K Ravikumar, and Eric P Xing. Dags with NO-TEARS: Continuous optimization for structure learning. Advances in Neural Information Processing Systems, 31, 2018.
There are two separate environments that need to be configured to reproduce this project: CDML and DECI (causica).
You may create the virtual environments with their respective requirements using the provided .yml files, using for example your Anaconda installation, on your shell as
-
For CDML:
conda env create -f environment-cdml.yml
-
For causica:
conda env create -f environment-causica.yml
- The first environment runs on Python 3.8.19.
- The second environment runs on Python 3.10.1, 🔥 PyTorch 1.13.0 and PyTorch lightning 2.2.2.
-
Run
generate_dataset.ipynbto generate a CDML configuration, plot the causal graph and generate the corresponding time-lagged dataset. -
Run
experiments.ipynbto run a DECI model on a CDML configuration, compute the metrics and compare to the ground truth graph, as well as PCMCI. -
Run
RunAll.ipynbto evaluate all pre-trained DECI models on each datasetavailable at thedatasetsfolder and compare with PCMCI.
Enjoy! 🚀