Your all-inclusive package for aggregating and visualizing metagenomic BLAST results.
Note that the pygraphviz python dependency has the graphviz non-python dependency. How to install it depends on your system. See the pygraphviz docs for details.
Here are a few common methods:
# conda
$ conda install pygraphviz
# Ubuntu and Debian
$ sudo apt-get install graphviz graphviz-dev
# Fedora and Red Hat
$ sudo dnf install graphviz graphviz-devel
# macOS
$ brew install graphvizAfterwards, metagenompy can be installed using pip:
$ pip install metagenompyThe core of metagenompy is a taxonomy as a networkX object.
This means that all your favorite algorithms work right out of the box.
import metagenompy
import networkx as nx
# load taxonomy
graph = metagenompy.generate_taxonomy_network(auto_download=True)
# print path from human to pineapple
for node in nx.shortest_path(graph.to_undirected(as_view=True), '9606', '4615'):
print(node, graph.nodes[node])
## 9606 {'rank': 'species', 'authority': 'Homo sapiens Linnaeus, 1758', 'scientific_name': 'Homo sapiens', 'genbank_common_name': 'human', 'common_name': 'man'}
## 9605 {'rank': 'genus', 'authority': 'Homo Linnaeus, 1758', 'scientific_name': 'Homo', 'common_name': 'humans'}
## [..]
## 4614 {'rank': 'genus', 'authority': 'Ananas Mill., 1754', 'scientific_name': 'Ananas'}
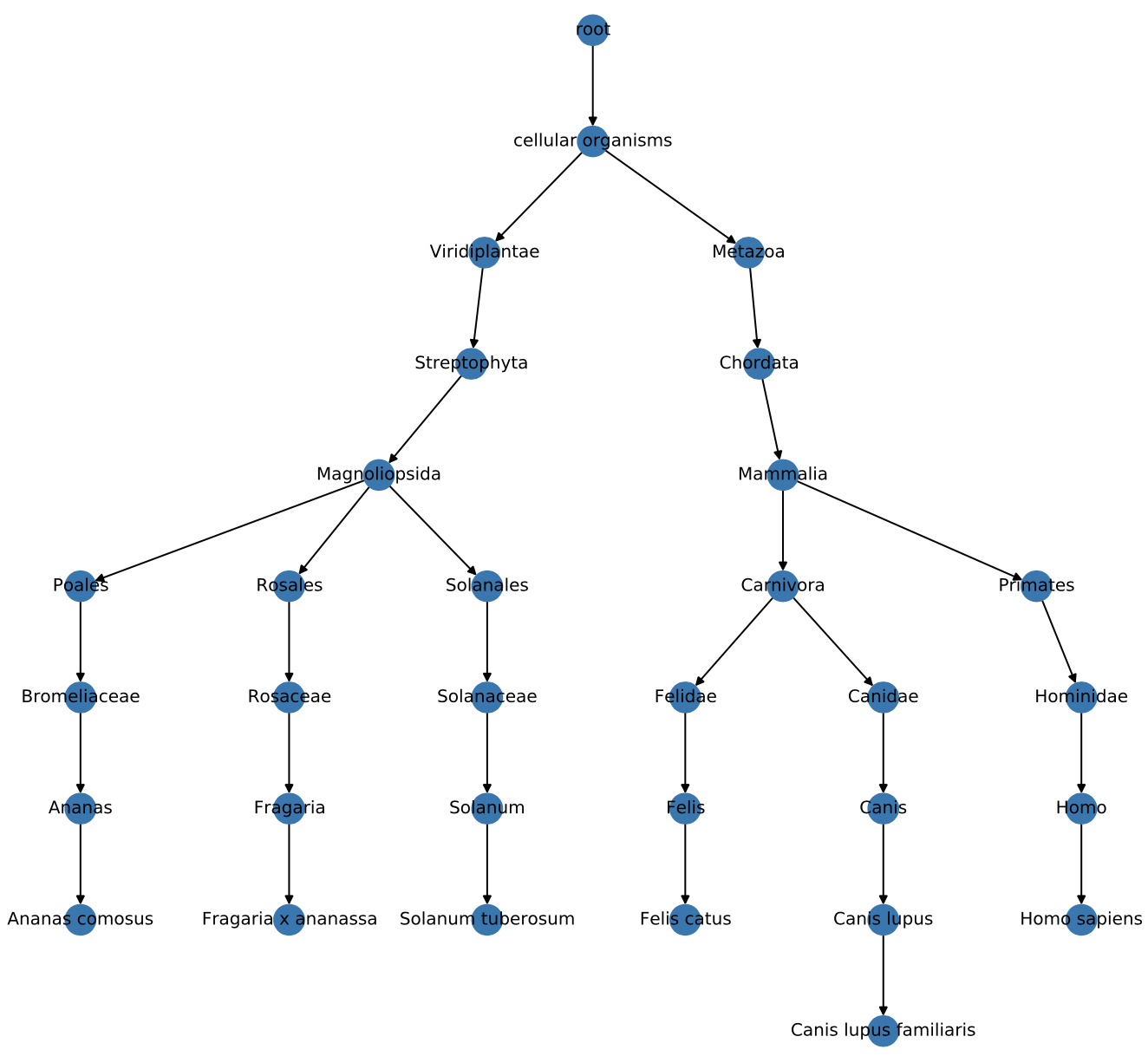
## 4615 {'rank': 'species', 'authority': ['Ananas comosus (L.) Merr., 1917', 'Ananas lucidus Mill., 1754'], 'scientific_name': 'Ananas comosus', 'synonym': ['Ananas comosus var. comosus', 'Ananas lucidus'], 'genbank_common_name': 'pineapple'}Extract taxonomic entities of interest and visualize their relations:
import metagenompy
import matplotlib.pyplot as plt
# load and condense taxonomy to relevant ranks
graph = metagenompy.generate_taxonomy_network(auto_download=True)
metagenompy.condense_taxonomy(graph)
# highlight interesting nodes
graph_zoom = metagenompy.highlight_nodes(graph, [
'9606', # human
'9685', # cat
'9615', # dog
'4615', # pineapple
'3747', # strawberry
'4113', # potato
])
# visualize result
fig, ax = plt.subplots(figsize=(10, 10))
metagenompy.plot_network(graph_zoom, ax=ax, labels_kws=dict(font_size=10))
fig.tight_layout()
fig.savefig('taxonomy.pdf')After blasting your reads against a sequence database, generating summary reports using metagenompy is a blast.
import metagenompy
import pandas as pd
# read BLAST results file with columns 'qseqid' and 'staxids'
df_blast = metagenompy.load_example_dataset()
df = (df_blast.set_index('qseqid')['staxids']
.str.split(';')
.explode()
.dropna()
.reset_index()
.rename(columns={'staxids': 'taxid'})
)
df.head()
## qseqid taxid
## 0 read1 1811693
## 1 read2 327160
## 2 read3 821
## 3 read4 1871047
## 4 read5 69360
# classify taxons at multiple ranks
graph = metagenompy.generate_taxonomy_network(auto_download=True)
rank_list = ['species', 'genus', 'class', 'superkingdom']
df = metagenompy.classify_dataframe(
graph, df,
rank_list=rank_list
)
# aggregate read matches
agg_rank = 'genus'
df_agg = metagenompy.aggregate_classifications(df, agg_rank)
df_agg.head()
## taxid species genus class superkingdom
## qseqid
## read1 1811693 Pelotomaculum sp. PtaB.Bin104 Pelotomaculum Clostridia Bacteria
## read10 2488860 Erythrobacter spongiae Erythrobacter Alphaproteobacteria Bacteria
## read100 78398 Pectobacterium odoriferum Pectobacterium Gammaproteobacteria Bacteria
## read101 1843082 Macromonas sp. BK-30 Macromonas Betaproteobacteria Bacteria
## read102 2665644 Paracoccus sp. YIM 132242 Paracoccus Alphaproteobacteria Bacteria
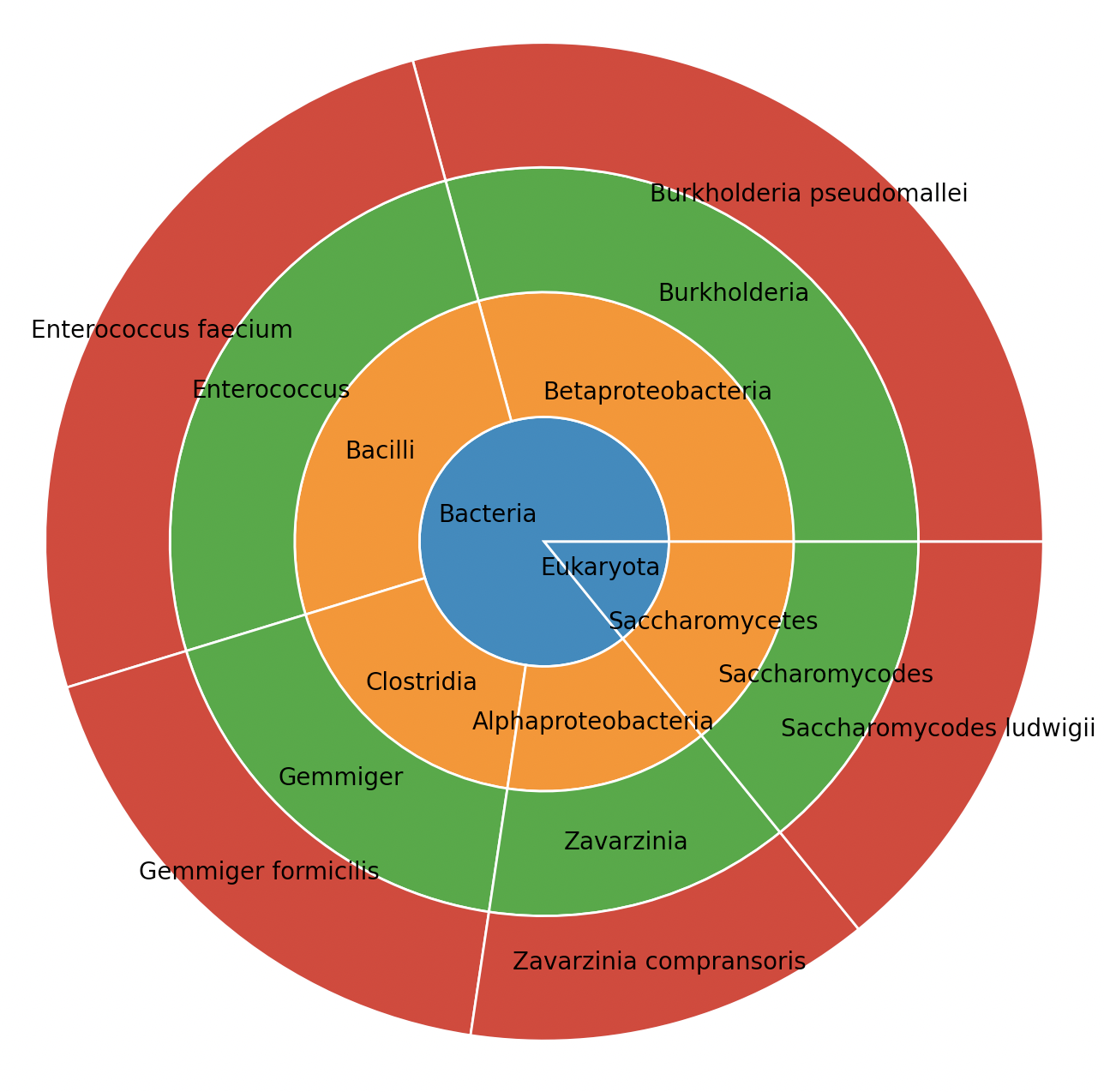
# visualize outcome
metagenompy.plot_piechart(df_agg)