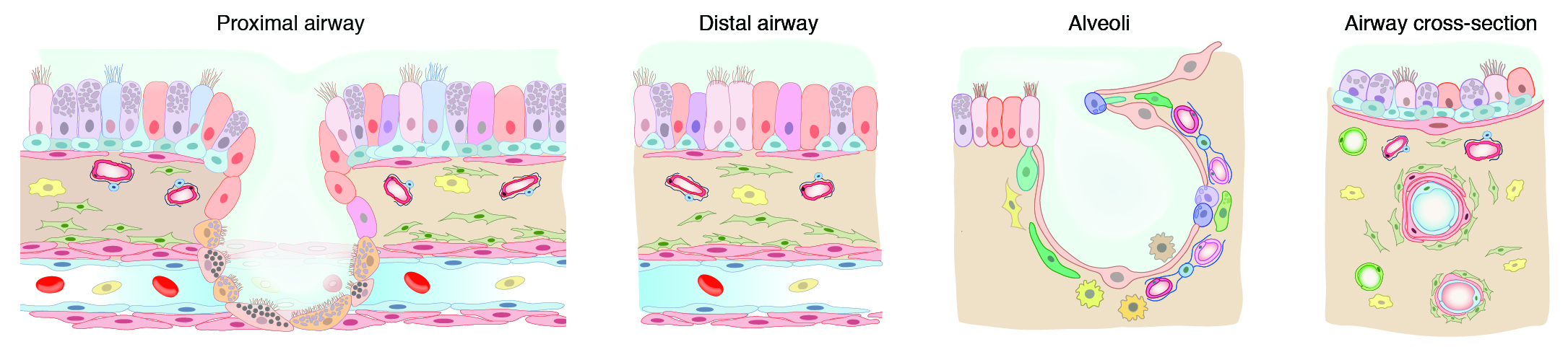
These R markdown notebooks import patient-specific gene count/UMI tables (SS2/10x) with Seurat, seperate the cells by tissue compartment, iteratively subclsuter them, and then assign each biologically meaningful cluster an identity based on canonical marker genes, novel bulk RNAseq markers (immune cells), tissue location, and biochemical function. Note: The specific version of Seurat used (as well as its dependencies) can produce slightly different clustering results. Early steps in these notebooks remove cells from patient-matched diseased regions of the lung, which are not yet released.
These notebooks import annotated, patient-specific Seurat objects (see Data availability below) from the annotation notebooks and merges them for downstream analyses. They explore the biochemical functions of lung cell types and the cell-selective transcription factors and optimal markers for making and monitoring them; define the cell targets of circulating hormones and predicts local signaling interactions including sources and targets of chemokines in immune cell trafficking and expression changes on lung homing; and identify the cell types directly affected by lung disease genes. They also compare human and mouse cell types to identify cell types whose expression profiles have been substantially altered by evolution, revealing extensive plasticity of cell-type-specific gene expression. These notebooks output the Figures and Tables used in the manuscript.
There are several files used by the Analysis notebooks that are too big for GitHub. To obtain them:
(1) Clone the GitHub repository
git clone http://github.com/krasnowlab/hlca
(2) Setup the Synapse CLI
(3) Navigate to the repository
(4) Run the download script
bash fetchData.bash
We reannotated two existing human lung single cell RNAseq datasets from the Teichmann and Misharin labs using our approach. Notebooks and CSVs containing the new annotations are provided.
Download count/UMI tables, metadata, and Seurat and scanpy objects for your own bioinformatic piplines from Synapse. Sequencing reads are available on the European Genome-phenome Archive. Please complete our Data Access Agreement and return to our Data Access Committee to request access.