This repository contains the R code for Buyukozkan*, Alvarez-Mulett*, Racanelli* et al., Integrative Metabolomic and Proteomic Signatures Define Clinical Outcomes in Severe COVID-19. https://doi.org/10.1101/2021.07.19.21260776
Each folder contains the codes for one analysis block of the paper:
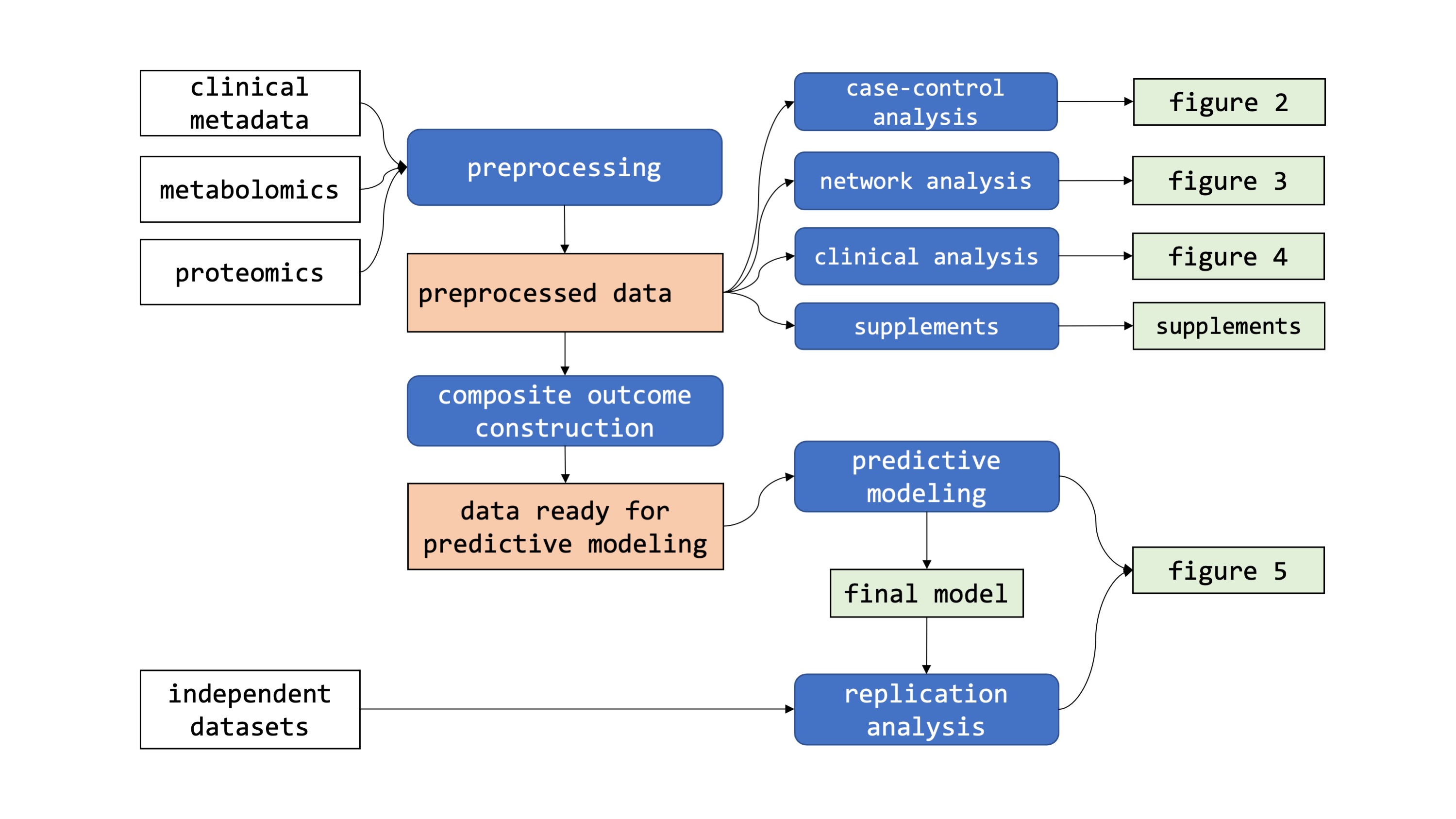
- 1_preprocessing - Preprocessing of raw metabolomics/proteomics data
- 2a_casecontrol - Differential metabolomics/proteomics analysis of COVID-19 patients vs. controls
- 2b_clinical - Differential metabolomics/proteomics analysis of clinical parameters within COVID-19 patients
- 2c_networks - GGM network analysis
- 2d_supplements - Analysis of intubation effects, time effects, and PCAs
- 3a_composite - Composite outcome construction
- 3b_prediction - Regularized linear mixed effect ordinal regression model
- 4_replication - Model testing on independent datasets
- 5_paperfigures - Generation of paper figures
Download the study data from https://doi.org/10.6084/m9.figshare.19115972.v1. Clone this git repository and copy all data files into a "DATA/" subdirectory in the git folder.
Note: During the first run, the main script will use the renv package to restore the correct versions of all required packages. This does not affect the system-wide library versions; all changes are stored in the local project.
main.R: runs the entire pipeline by sourcing the all scripts listed below
run1_preprocessing.R: runs all data preprocessing stepsrun2_allanalysis.R: runs statistical association analysis and network analysisrun3_prediction.R: runs predictive modelingrun4_replication.R: runs replication analysisrun5_figures.R: generates paper figures
See expected directory tree after running all the code.