The 'Regularized PAIrwise Ranking', or rpair, package is a convenient toolbox for conducting analyses on high-dimensional omics data. rpair combines a general framework for casting complex outcomes into pairwise learning problems with a penalty function allowing for sparse solutions. This results in models that are not only able to fully utilize such data, but are far more interpretable than traditional methods such as the Cox proportional hazard model. The package supports four types of loss functions: logistic loss, exponential loss, squared hinge loss and huberized hinge loss. [JK: intro needs to be adapted, up to this point it could be glmnet. currently discussed in slack]
Buyukozkan, Chetnik, and Krumsiek. "Regularized PAIrwise Ranking survival analysis: RPAIR". (unpublished) 2022
The rpair package can be installed using the following command:
require(devtools)
devtools::install_github(repo="krumsieklab/rpair", subdir="rpair")# libraries
library(rpair)
library(magrittr)
library(survival)
# load example data
x <- rpair::ds1_x
y <- rpair::ds1_y
# run cross-validated rpair
set.seed(44)
cv = cv_rpair(x, y)
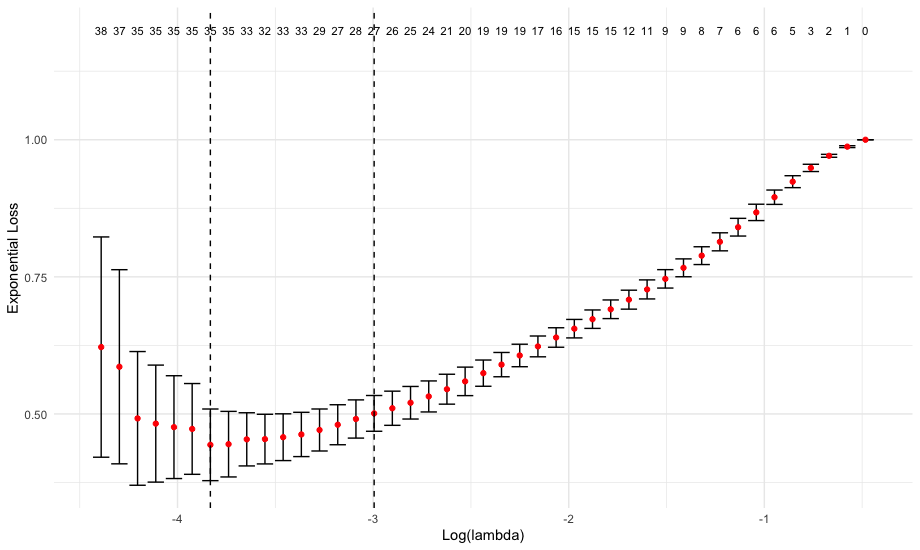
# lambda plot
plot(cv)
# apply model to input data
pr = predict(cv, x)
# show predicted survival times
pr[1:10][1] 1.8386803 -2.7819668 -0.9915281 -1.2221169 -0.5459928 0.2180884 -0.6377641 2.6466038 1.8887652 -2.5777407
Detailed examples illustrating the full functionality of the package are provided in the following tutorials: