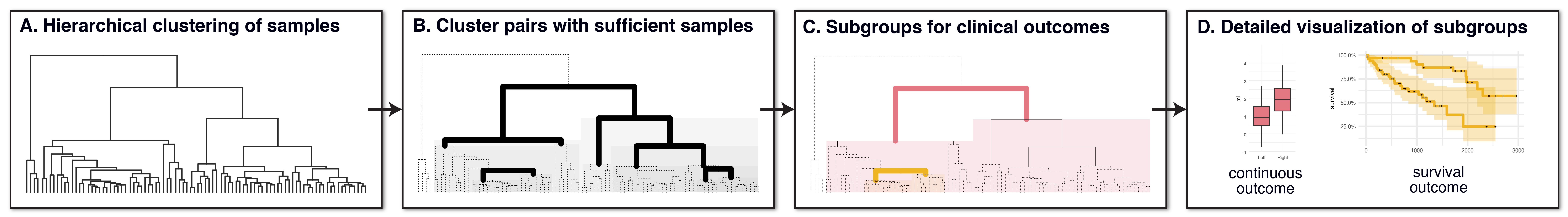
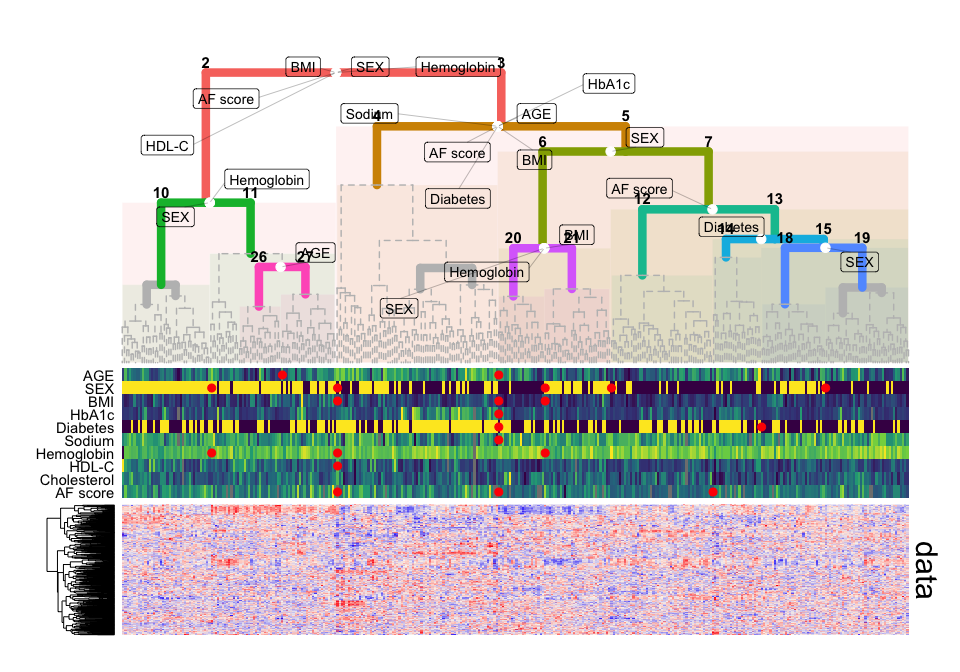
The 'Subgroup Identification' (SGI) toolbox provides an algorithm to
automatically detect clinical subgroups of samples in large-scale omics
datasets. It is based on hierarchical clustering trees in combination
with a specifically designed association testing and visualization
framework that can process a large number of clinical parameters and
outcomes in a systematic fashion. A multi-block extension allows for the
simultaneous use of multiple omics datasets on the same samples.
Buyukozkan, et al. "SGI: Automatic clinical subgroup identification in omics datasets". Bioinformatics, 2021. link to publication
SGI can be installed as follows:
require(devtools)
devtools::install_github(repo="krumsieklab/sgi", subdir="sgi")Here are a few lines of code that demostrate how SGI works:
library(sgi)
# hierarchical clustering
hc = hclust(dist(sgi::qmdiab_plasma), method = "ward.D2")
# initialize SGI structure; minsize is set to 5% of sample size
sg = sgi_init(hc, minsize = 18, outcomes = sgi::qmdiab_clin)
# run SGI
as = sgi_run(sg)
# generate tree plot, show results for adjusted p-values <0.05
gg_tree = plot(as, padj_th = 0.05)
# plot overview, including clinical data and metabolomics data matrix
plot_overview( gg_tree = gg_tree, as = as,
outcomes = sgi::qmdiab_clin,
xdata = sgi::qmdiab_plasma )For more detailed examples and functionalities of the package, we provide the following tutorials:
-
SGI Tutorial 1: Quickstart guide provides a quick introduction to the functionality of the SGI package.
-
SGI tutorial 2: Objects and functions explains the functionality of SGI objects for result extraction and plotting.
-
SGI tutorial 3: Detailed examples of plotting functionality showcases various plotting scenarios for SGI results.
-
SGI tutorial 4: Multi-omics SGI showcases the multi-omics capabilities of SGI
-
SGI tutorial 5: SGI with user-defined statistical tests explains how user-defined tests can be used in SGI