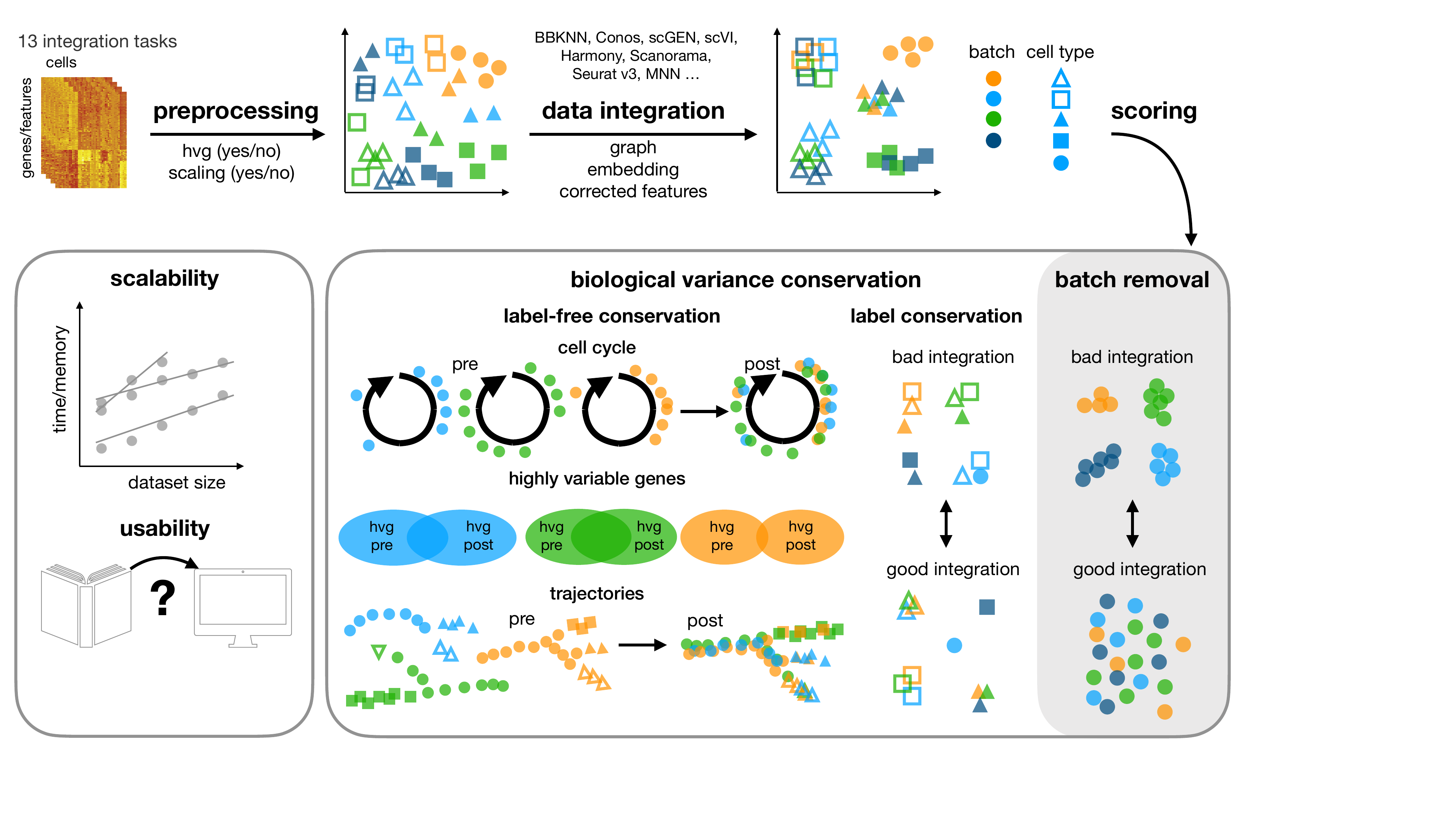
This repository contains the snakemake pipeline for our benchmarking study for data integration tools.
In this study, we benchmark 16 methods (see here) with 4 combinations of preprocessing steps leading to 68
methods combinations on 85 batches of gene expression and chromatin accessibility data.
The pipeline uses the scIB package and allows for reproducible and automated
analysis of the different steps and combinations of preprocesssing and integration methods.
-
On our website we visualise the results of the study.
-
The scib package that is used in this pipeline can be found here.
-
For reproducibility and visualisation we have a dedicated repository: scib-reproducibility.
_Benchmarking atlas-level data integration in single-cell genomics.
MD Luecken, M Büttner, K Chaichoompu, A Danese, M Interlandi, MF Mueller, DC Strobl, L Zappia, M Dugas, M Colomé-Tatché, FJ Theis
bioRxiv 2020.05.22.111161; doi: https://doi.org/10.1101/2020.05.22.111161 _
To reproduce the results from this study, three different conda environments are needed. There are different environments for the python integration methods, the R integration methods and the conversion of R data types to anndata objects.
The main steps are:
- Install the conda environment
- Set environment variables
- Install any extra packages through
R
For the installation of conda, follow these instructions or use your system's package manager. The environments have only been tested on linux operating systems although it should be possible to run the pipeline using Mac OS.
To create the conda environments use the .yml files in the envs directory.
To install the envs, use
conda env create -f FILENAME.ymlNote: Instead of
condayou can usemambato speed up installation times
For R environments, some dependencies need to be installed after the environment has been created. However, it is important to set environment variables for the conda environments first, to guarantee that the correct R version installs packages into the correct directories. All necessary steps are mentioned below.
Some parameters need to be added manually to the conda environment in order for packages to work correctly.
For example, all environments using R need LD_LIBRARY_PATH set to the conda R library path.
If that variable is not set, rpy2 might reference the library path of a different R installation that might be on your system.
Environment variables are provided in env_vars_activate.sh and env_vars_deactivate.sh and should be copied to the designated locations of each conda environment.
Make sure to determine $CONDA_PREFIX in the activated environment first, then deactivate the environment before copying the files to prevent unwanted effects.
This process is automated with the following script, which you should call for each environment that uses R.
. envs/set_vars.sh <conda_prefix>After the script has successfully finished, you should be ready to use your new environment.
If you want to set these and potentially other variables manually, proceed as follows.
e.g. for scIB-python:
conda activate scIB-python
echo $CONDA_PREFIX # referred to as <conda_prefix>
conda deactivate
# copy activate variables
cp envs/env_vars_activate.sh <conda_prefix>/etc/conda/activate.d/env_vars.sh
# copy deactivate variables
cp envs/env_vars_deactivate.sh <conda_prefix>/etc/conda/deactivate.d/env_vars.shIf necessary, create any missing directories manually. In case some lines in the environment scripts cause problems, you can edit the files to trouble-shoot.
There are multiple different environments for the python dependencies:
| YAML file location | Environment name | Description |
|---|---|---|
envs/scib-pipeline.yml |
scib-pipeline |
Base environment for calling the pipeline, running python integration methods and computing metrics |
envs/scIB-python-paper.yml |
scIB-python-paper |
Environment used for the results in the [publication](doi: https://doi.org/10.1101/2020.05.22.111161) |
The scib-pipeline environment is the one that the user activates before calling the pipeline.
It needs to be specified under the py_env key in the config files under configs/ so that the pipeline will use it for running python methods. Alternatively, you can specify scIB-python-paper as the py_env to recreate the environment used in the paper to reproduce the results.
Furthermore, scib-pipeline python environments require the R package kBET to be installed manually. This also requires that environment variables are set as described above, so that R packages are correctly installed and located. Once environment variables have been set, you can install kBET:
conda activate <py-environment>
Rscript -e "devtools::install_github('theislab/kBET')"
| YAML file location | Environment name | Description |
|---|---|---|
envs/scIB-R-integration.yml |
scIB-R-integration |
Environment used for the results in the [publication](doi: https://doi.org/10.1101/2020.05.22.111161) |
envs/scib-R.yml |
scib-R |
Updated environment with R dependencies |
The R environments require extra R packages to be installed manually.
Activate the environment and install the packages all the R dependencies in R directly or use the script install_R_methods.R.
conda activate <r-environment>
Rscript envs/install_R_methods.R
For the installation ofConos, please see the Conos github repo.
We used these conda versions of the R integration methods in our study:
harmony_1.0
Seurat_3.2.0
conos_1.3.0
liger_0.5.0
batchelor_1.4.0
This repository contains a snakemake pipeline to run integration methods and metrics reproducibly for different data scenarios preprocessing setups.
A script in data/ can be used to generate test data.
This is useful, in order to ensure that the installation was successful before moving on to a larger dataset.
More information on how to use the data generation script can be found in data/README.md.
The parameters and input files are specified in config files, that can be found in configs/.
In the DATA_SCENARIOS section you can define the input data per scenario.
The main input per scenario is a preprocessed .h5ad file of an anndata with batch and cell type annotations.
TODO: explain different entries
To call the pipeline on the test data
snakemake --configfile configs/test_data.yaml -n
This gives you an overview of the jobs that will be run. In order to execute these jobs, call
snakemake --configfile configs/test_data.yaml --cores N_CORES
where N_CORES defines the number of threads to use.
More snakemake commands can be found in the documentation.
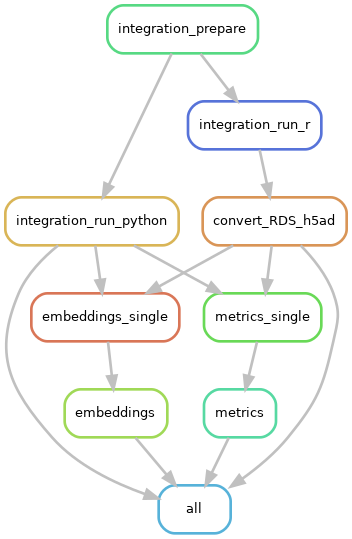
A dependency graph of the workflow can be created anytime and is useful to gain a general understanding of the workflow.
Snakemake can create a graphviz representation of the rules, which can be piped into an image file.
snakemake --configfile configs/test_data.yaml --rulegraph | dot -Tpng -Grankdir=TB > dependency.pngTools that are compared include: