Projekt KWD
Jakub Pranica Robert Trojan
Identyfikacja choroby Parkinsona na podstawie mowy (Parkinson’s Data Set) - Neural network for binary classficiation
Zbudowaliśmy sieć neuronową z 5 warstwami mającymi różne funkcje aktywacji. Model wytrenowaliśmy i zwizualizowaliśmy wyniki w sposób przedstawiony poniżej.
Opis zestawu danych: Parkinsons Disease
Oxford Parkinson's Disease Detection Dataset
This dataset is composed of a range of biomedical voice measurements from 31 people, 23 with Parkinson's disease (PD). Each column in the table is a particular voice measure, and each row corresponds one of 195 voice recording from these individuals ("name" column). The main aim of the data is to discriminate healthy people from those with PD, according to "status" column which is set to 0 for healthy and 1 for PD.
The data is in ASCII CSV format. The rows of the CSV file contain an instance corresponding to one voice recording. There are around six recordings per patient, the name of the patient is identified in the first column.For further information or to pass on comments, please contact Max Little (littlem '@' robots.ox.ac.uk).
Attribute Information:
Matrix column entries (attributes):
- name - ASCII subject name and recording number
- MDVP:Fo(Hz) - Average vocal fundamental frequency
- MDVP:Fhi(Hz) - Maximum vocal fundamental frequency
- MDVP:Flo(Hz) - Minimum vocal fundamental frequency
- MDVP:Jitter(%),MDVP:Jitter(Abs),MDVP:RAP,MDVP:PPQ,Jitter:DDP - Several measures of variation in fundamental frequency
- MDVP:Shimmer,MDVP:Shimmer(dB),Shimmer:APQ3,Shimmer:APQ5,MDVP:APQ,Shimmer:DDA - Several measures of variation in amplitude
- NHR,HNR - Two measures of ratio of noise to tonal components in the voice
- status - Health status of the subject (one) - Parkinson's, (zero) - healthy
- RPDE,D2 - Two nonlinear dynamical complexity measures
- DFA - Signal fractal scaling exponent
- spread1,spread2,PPE - Three nonlinear measures of fundamental frequency variation
dataset: https://archive.ics.uci.edu/ml/machine-learning-databases/parkinsons/parkinsons.data
Importy
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import confusion_matrix, accuracy_score
from keras.layers import Dropout
#set seed for reproduction purpose
from numpy.random import seed
seed(1)
from tensorflow import set_random_seed
set_random_seed(2)
import random as rn
rn.seed(12345)
import tensorflow as tf
tf.set_random_seed(1234)
#import seaborn as snsWczytanie i standaryzacja danych
url = "https://archive.ics.uci.edu/ml/machine-learning-databases/parkinsons/parkinsons.data"
names = ["name","MDVP:Fo(Hz)","MDVP:Fhi(Hz)","MDVP:Flo(Hz)","MDVP:Jitter(%)","MDVP:Jitter(Abs)","MDVP:RAP","MDVP:PPQ",
"Jitter:DDP","MDVP:Shimmer","MDVP:Shimmer(dB)","Shimmer:APQ3","Shimmer:APQ5","MDVP:APQ","Shimmer:DDA",
"NHR","HNR","status","RPDE","DFA","spread1","spread2","D2","PPE"]
parkinson_df = pd.read_csv(url, names=names) #load CVS data
#load Pandas Dataframe into numpy arrays
data = parkinson_df.loc[1:,["MDVP:Fo(Hz)","MDVP:Fhi(Hz)","MDVP:Flo(Hz)","MDVP:Jitter(%)","MDVP:Jitter(Abs)","MDVP:RAP","MDVP:PPQ",
"Jitter:DDP","MDVP:Shimmer","MDVP:Shimmer(dB)","Shimmer:APQ3","Shimmer:APQ5","MDVP:APQ","Shimmer:DDA",
"NHR","HNR","RPDE","DFA","spread1","spread2","D2","PPE"]].values.astype(np.float)
target = parkinson_df.loc[1:, ['status']].values.astype(np.float)
#standarise data
data = StandardScaler().fit_transform(data)
data_train, data_test, target_train, target_test = \
train_test_split(data, target, test_size=0.3, random_state=545)Budowa sieci neuronowej za pomocą Kerasa - Model sekwencyjny
from keras.models import Sequential
from keras.layers import Dense
from keras.optimizers import SGD
neural_model = Sequential([
Dense(16, input_shape=(22,), activation="relu"),
Dense(8, activation="exponential"),
Dense(4, activation='tanh'),
Dense(2, activation='linear'),
Dense(1, activation="sigmoid")
])
#show summary of a model
neural_model.summary()_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_88 (Dense) (None, 16) 368
_________________________________________________________________
dense_89 (Dense) (None, 8) 136
_________________________________________________________________
dense_90 (Dense) (None, 4) 36
_________________________________________________________________
dense_91 (Dense) (None, 2) 10
_________________________________________________________________
dense_92 (Dense) (None, 1) 3
=================================================================
Total params: 553
Trainable params: 553
Non-trainable params: 0
_________________________________________________________________
Trenowanie modelu
neural_model.compile(SGD(lr = .003), "binary_crossentropy", \
metrics=["accuracy"])
np.random.seed(0)
run_hist_1 = neural_model.fit(data_train, target_train, epochs=4000,\
validation_data=(data_test, target_test), \
verbose=False, shuffle=False)
print("Training neural network...\n")
print('Accuracy over training data is ', \
accuracy_score(target_train, neural_model.predict_classes(data_train)))
print('Accuracy over testing data is ', \
accuracy_score(target_test, neural_model.predict_classes(data_test)))
conf_matrix = confusion_matrix(target_test, neural_model.predict_classes(data_test))
print(conf_matrix)Training neural network...
Accuracy over training data is 1.0
Accuracy over testing data is 0.8813559322033898
[[10 5]
[ 2 42]]
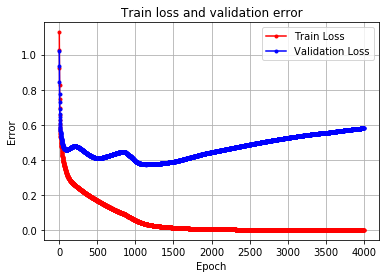
Wizualizacja procesu uczenia
#run_hist_1.history.keys()
plt.plot(run_hist_1.history["loss"],'r', marker='.', label="Train Loss")
plt.plot(run_hist_1.history["val_loss"],'b', marker='.', label="Validation Loss")
plt.title("Train loss and validation error")
plt.legend()
plt.xlabel('Epoch'), plt.ylabel('Error')
plt.grid()Można zauważyć przyuczenie do danych uczących. Dodaliśmy więc warstwy Dropout, co pozwoliło uzyskać lepsze wyniki dla zestawu walidacyjnego.
Budowa modelu z Dropoutami
#model with dropouts
neural_network_d = Sequential()
neural_network_d.add(Dense(16, activation='relu', input_shape=(22,)))
neural_network_d.add(Dense(8, activation="exponential"))
neural_network_d.add(Dropout(0.1))
neural_network_d.add(Dense(4, activation='tanh'))
neural_network_d.add(Dense(2, activation='linear'))
neural_network_d.add(Dropout(0.1))
neural_network_d.add(Dense(1, activation='sigmoid'))
neural_network_d.summary()_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_98 (Dense) (None, 16) 368
_________________________________________________________________
dense_99 (Dense) (None, 8) 136
_________________________________________________________________
dropout_28 (Dropout) (None, 8) 0
_________________________________________________________________
dense_100 (Dense) (None, 4) 36
_________________________________________________________________
dense_101 (Dense) (None, 2) 10
_________________________________________________________________
dropout_29 (Dropout) (None, 2) 0
_________________________________________________________________
dense_102 (Dense) (None, 1) 3
=================================================================
Total params: 553
Trainable params: 553
Non-trainable params: 0
_________________________________________________________________
neural_network_d.compile(SGD(lr = .003), "binary_crossentropy", metrics=["accuracy"])
run_hist_2 = neural_network_d.fit(data_train, target_train, epochs=4000, \
validation_data=(data_test, target_test), \
verbose=False, shuffle=False)
print("Training neural network w dropouts..\n")
print('Accuracy over training data is ', accuracy_score(target_train, \
neural_network_d.predict_classes(data_train)))
print('Accuracy over testing data is ', accuracy_score(target_test, \
neural_network_d.predict_classes(data_test)))
Training neural network w dropouts..
Accuracy over training data is 0.9926470588235294
Accuracy over testing data is 0.9322033898305084
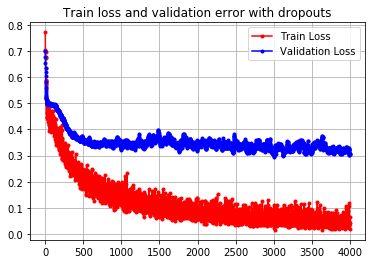
plt.plot(run_hist_2.history["loss"],'r', marker='.', label="Train Loss")
plt.plot(run_hist_2.history["val_loss"],'b', marker='.', label="Validation Loss")
plt.title("Train loss and validation error with dropouts")
plt.legend()
plt.grid()Dropouty pomagają uniknąć zbyt dużego przyuczenia do danych.