Code and reproducible example for computing the Taylor, solar and target diagrams.
Based on the paper: Wadoux A.M.J-C., Walvoort D.J.J and Brus D.J. (2021) An integrated approach for the evaluation of quantitative soil maps through Taylor and solar diagrams (Geoderma)
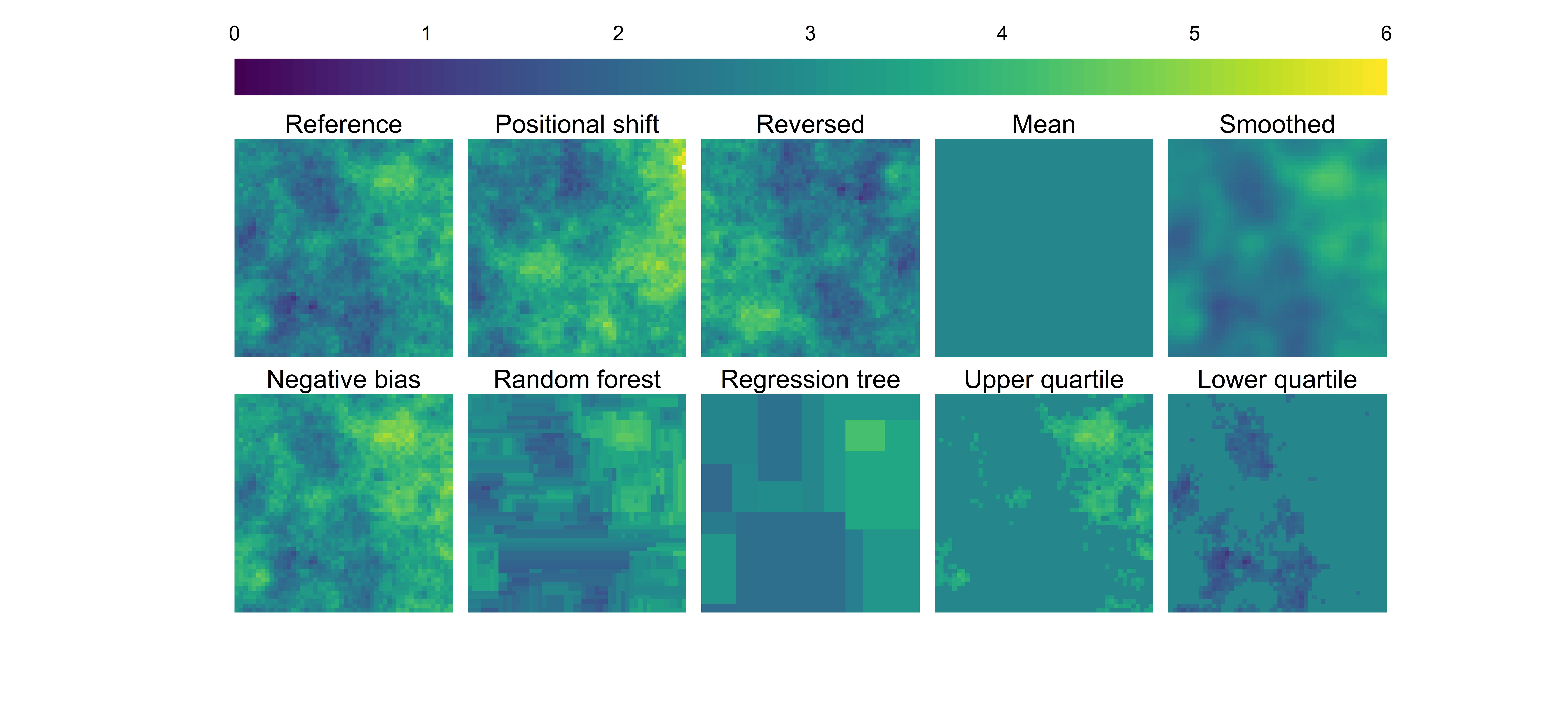
To reproduce the figures below, run the file: "Example_paper.R"- grid of size 50 × 50 cells
- we simulate a realisation of a map composed of a linear spatial trend superimposed on a Gaussian random field
- trend has an intercept of 5 and slope parameter of 0.1 for the x-axis and 0.05 for the y-axis
- the Gaussian random field has a mean of zero and a covariance given by C(h) = 5 exp(-h/10), where h is the lag distance
- this map is multiplied by a factor of 0.3 to obtain the ultimate reference map
- a map positionally shifted by 20 units in the x-direction and the y-direction,
- a map where the x- and y-coordinates are reversed,
- a map containing the mean value of the reference map at each coordinate,
- a smoothed map obtained by a moving average window of size 5 x 5 units,
- a negatively biased map obtained by adding a value of 1 to the reference map,
- a map predicted by a random forest model with a single tree and default parameters from the R package ranger using the reference map as calibration data and the x- and y-coordinates as predictors,
- a map predicted by a single regression tree with default parameters from the R package rpart using the reference map as calibration data and the x- and y-coordinates as predictors,
- a map of the upper quartile, made by assigning the mean of the reference map to the values lower than the upper quartile and value of the reference map otherwise,
- a map of the lower quartile, made by assigning the mean of the reference map to the values higher than the lower quartile and the value of the reference map otherwise.
# load function
source('./R_Taylor_function.R')
# make Target diagram
gg_taylor(mods = models,
obs = obser,
label = TRUE)# load function
source('./R_solar_function.R')
# make solar diagram
gg_solar(mods = models,
obs = obser,
colorval = colorval,
colorval.name = 'MEC',
label = TRUE,
x.axis_begin = -1.1,
x.axis_end = 1.1,
y.axis_end = 1.9,
by = 0.1)# load function
source('./R_target_function.R')
# make target diagram
gg_target(mods = models,
obs = obser,
colorval = colorval,
colorval.name = 'MEC',
label = TRUE,
axis_begin = -2.2,
axis_end = 2.2,
by = 0.1)