Package written in Python 3.6.0, using Keras version 2.0.1 with a Tensflow (version 1.0.0) backend
Author: Lukmaan A. Bawazer
This project optimizes deep learning model architectures via genetic programming. Currently only simple fully connected architectures are supported, with plans to extend the package to RNN and CNN architectures in the near future.
In addition to the notes below, a step-by-step introduction is provided in the demos folder.
Currently the package is set up to be run interactively (e.g., from an Jupyter notebook), using sacred as a tool to edit a default config dictionary which defines parameters for an experiment. (In the future an automated evolution option will be added, whereby diversification, selection, and breed occur automatically across a specified number of model generations.)
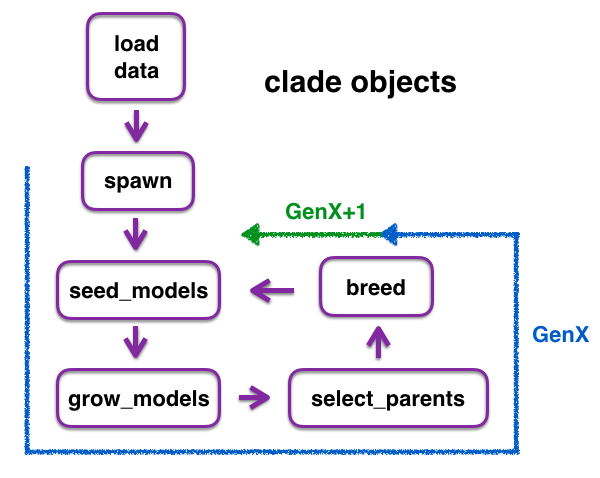
The default config dictionary values are defined in environment.py. These values specify hyperparameter ranges that are sampled to build a population of fully connected Tensorflow architectures, which are built using Keras. Each model in a population is trained and evaluated, and high-performing (and some random) sub-sets of models are selected as 'parents' to generate a new population of models based on the parameters of the selected parents. These operations are performed via a clade object, and can be iterated to conduct multiple generations of model evolution via the object. However, it is not necessary to use the same clade object for all steps in the evolution. Outputs of functions are saved as either serialized models (.h5 files) or pickled dataframes. If desired or useful, outputs from a function (e.g., dataframes with model test performances) of one clade object can be loaded into a downstream function (e.g, to pick top models in select_parents) in a different clade object.
The utility of deeplearngene is here demonstrated using the Reuters news articles dataset found in Keras. In the code posted here, this dataset is loaded into each clade object through the load_model() function, but this function can be edited to load any dataset of interest.
To work with deeplearngene interactively, first import the clades module, the sacred object from the environment module, and pandas:
import clades
from environment import ex
import pandas as pdTo use the default config dictionary, define a dictionary by running the sacred object.
config_dict = ex.run()Or make changes to the config dictionary via config_updates if desired.
config_dict = ex.run(config_updates={'population_size':80})Define a GAFC1 clade object. "GAFC1" stands for Genetic Algorithm operations for Fully Connected architectures version 1. Creation of additional clade objects, for use in evolving different types of deep learning architectures, is planned.
#.config refers to the config dictionary of the sacred object
my_clade = clades.GAFC1(config_dict.config)Clade functions are now available in my_clade, including:
- my_clade.load_data()
- Loads the data into the clade object
- my_clade.spawn()
- Creates "genes" that define specific model architectures.
Output: a pickled "genotypes.p" dataframe, which is also stored in the object as a property (my_clade.genotypes), where each row in the dataframe encodes a different keras fully connected model architecture
- Creates "genes" that define specific model architectures.
- my_clade.seed_models()
- Constructs, compiles, and saves (.h5 serializes) all keras models encoded in a genotypes dataframe
- my_clade.grow_models()
- Loads saved models, then trains and evaluates them. Uses the MonitorGrowth callback function (defined in callbacks.py) to track batch- and epoch-level performance.
Output:- A pickled "phenotypes.p" dataframe, which is also stored in the object as a property (my_clade.phenotypes). Each row in this dataframe summarizes the train and test loss and accuracy for each model, and also records the time taken to run each model.
- Pickled "growth_analysis.p" dataframes--one of each trained model. These dataframes include the train and validation loss and accuracy for each batch and epoch of training, and also the time taken in seconds to run each batch and epoch.
- Saved (.h5 serialized) trained keras models.
- Loads saved models, then trains and evaluates them. Uses the MonitorGrowth callback function (defined in callbacks.py) to track batch- and epoch-level performance.
- my_clade.select_parents()
- Uses a phenotypes dataframe to select a subsets of models based on performance metric thresholds (by default, the 20% best performing models by test accuracy, plus 10% models randomly selected from the phenotype dataframe).
Output: Stores selected models as an object property, my_clade.parent_genes.
- Uses a phenotypes dataframe to select a subsets of models based on performance metric thresholds (by default, the 20% best performing models by test accuracy, plus 10% models randomly selected from the phenotype dataframe).
- my_clade.breed()
- Produces a new genotypes dataframe by recombining encoded hyperparameters from selected parents and by randomly mutating hyperparamter values.
src/envrionment.py
Uses sacred to define default values of the config dictionary used for model diversification and selection.
src/clade.py
Defines the parent clade class, and takes care of data loading and some preprocessing (including train/validation/test splitting) in the load_data() class function.
src/clades.py
Defines clade sub-classes (currently just GAFC1) and implements most clade functions (spawn, breed, seed_models, grow_models, and select_parents).
src/callbacks.py
Defines callbacks (inheritance from the Keras Callback class) used during model training. The callback is called as models are trained in the clades.grow_models() function. Currently only one callback object, MonitorGrowth, is defined. MonitorGrowth includes an early stopping option, such that training will terminate at the end of an epoch if training has gone longer than a maximum specified time duration.
src/evaluations.py
Defines helper evaluation functions used to build the phenotypes dataframe as models are evaluated in clades.grow_models(). Currently only one helper function, onehot_misclassified(), is defined. This functino records each misclassified datapoint along with
the correct class assignment.
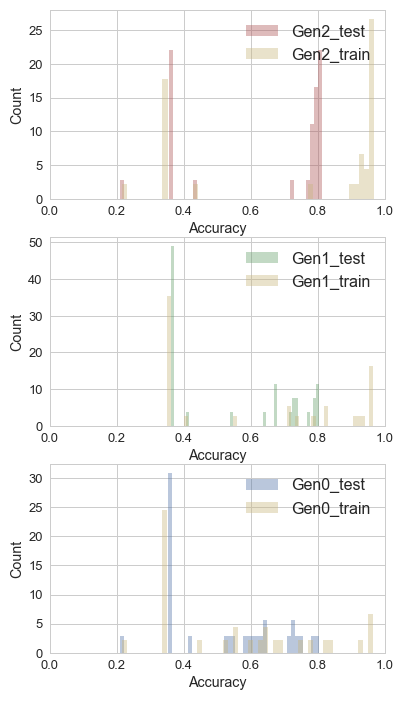
Below are histograms that summarize a demo experiment in which fully connected architectures were evolved over 3 generations (30 models per generation) to conduct NLP-based classification of Reuters news articles into 46 different news topics (see demos for more info). Each histogram plots the train and test accuracies of the 30 models from a given generation. The histograms show that model accuracies improve in the third generation, and the models suffer from over-fitting. The over-fitting issue can likely be addressed in future versions of deeplearngene by regularization, which will be added to the model diversification landscape.
- Extend package to include regularization (L1, L2, and Dropout)
- Extend to evolve RNN and CNN Architectures
- Extend to include more output metrics and visualizations
- Extend to run pools of models psuedo-distributed on virtual machines