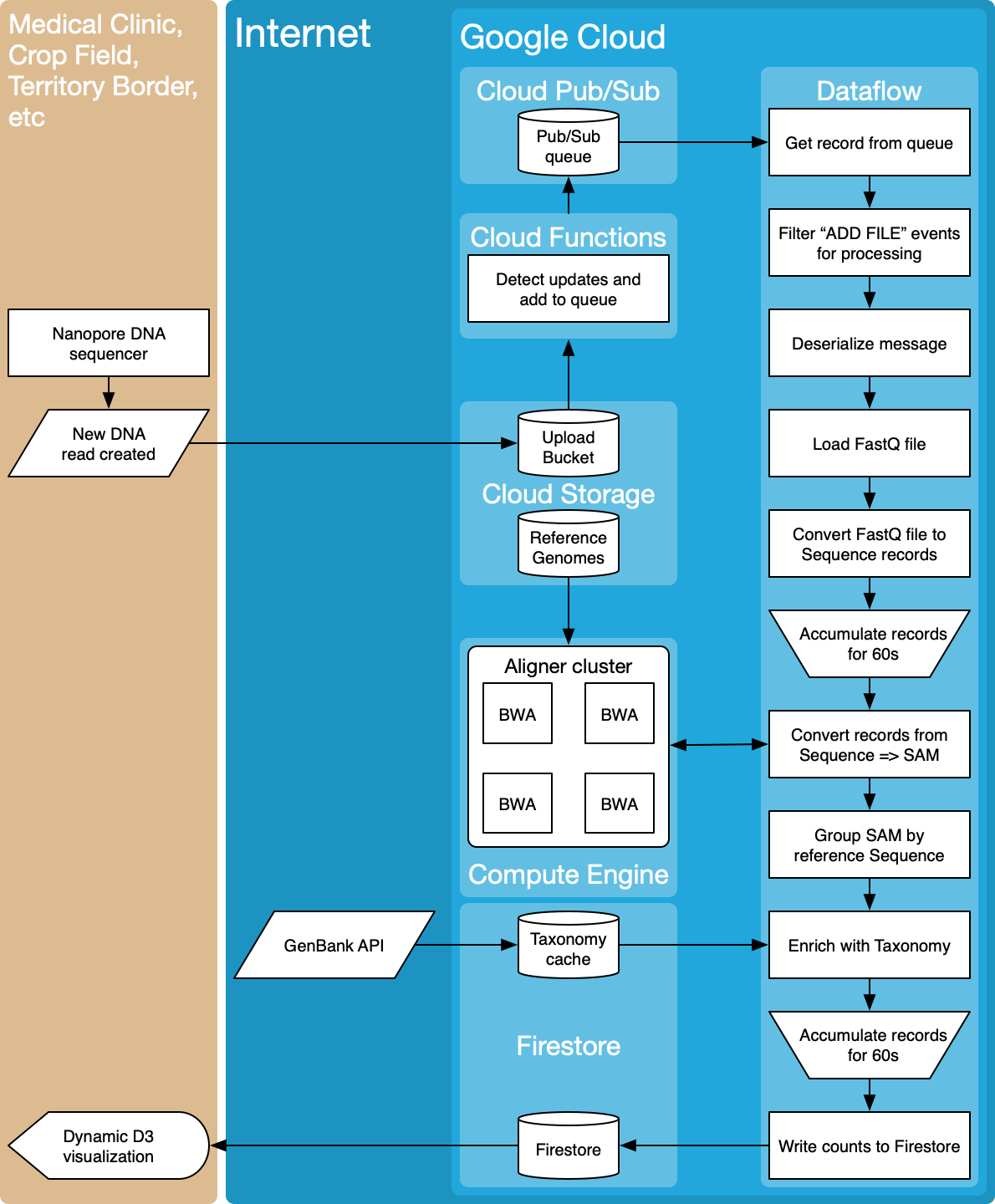
In a healthcare setting, being able to access data quickly is vital. For example, a sepsis patient’s survival rate decreases by 6% for every hour we fail to diagnose the species causing the infection and its antibiotic resistance profile.
Typical genomic analyses are too slow, taking weeks or months to complete. You transport DNA samples from the collection point to a centralized facility to be sequenced and analyzed in a batch process. Recently, nanopore DNA sequencers have become commercially available, such as those from Oxford Nanopore Technologies, streaming raw signal-level data as they are collected and providing immediate access to it. However, processing the data in real-time remains challenging, requiring substantial compute and storage resources, as well as a dedicated bioinformatician. Not only is the process is too slow, it’s also failure-prone, expensive, and doesn’t scale.
This source repo contains a prototype implementation of a scalable, reliable, and cost effective end-to-end pipeline for fast DNA sequence analysis using Dataflow on Google Cloud.
To run the pipeline take the following steps:
- Create a Google Cloud Project
- Create a Google Cloud Storage
$UPLOAD_BUCKETupload_bucket for FastQ files. - You can use our simulator to upload FastQ for testing, or if you don't have a real dataset.
- Configure file upload notifications. This creates PubSub messages when new files are uploaded. With our placeholder name
$UPLOAD_EVENTS, set up PubSub notifications in the following way:
gsutil notification create -t $UPLOAD_EVENTS -f json -e OBJECT_FINALIZE $UPLOAD_BUCKET
- Create a PubSub subscription for
$UPLOAD_EVENTS. With our placeholder name$UPLOAD_SUBSCRIPTION, run following command:
gcloud pubsub subscriptions create $UPLOAD_SUBSCRIPTION --topic $UPLOAD_EVENTS
- Provision an aligner cluster, see aligner
- Create a Firestore DB (See details) for saving cache and result data.
- optional If you running the pipeline in
resistance_genesmode you should provide "FASTA DB" and "gene list" files stored in GCS.
- NanostreamDataflowMain - Apache Beam app that provides all data transformations
- aligner - scripts to provision auto-scaled HTTP service for alignment (based on
bwa) - simulator - python script that can simulate file uploads to GCS
- fasta_formatter - python script for formatting fasta files into project readable format
- visualization - module for the visualization of results
- doc - additional files for documentation
We provide a pre-built jar file. See below if you want to build the jar yourself.
To run the pipeline, first define variables for configuration:
# Google Cloud project name
PROJECT=`gcloud config get-value project`
# Apache Beam Runner (Dataflow for Google Cloud Dataflow running or Direct for local running)
RUNNER=org.apache.beam.runners.dataflow.DataflowRunner
# specify mode of data processing (species, resistance_genes)
PROCESSING_MODE=resistance_genes
# PubSub subscription defined above
UPLOAD_SUBSCRIPTION=$UPLOAD_SUBSCRIPTION
# size of the window (in wallclock seconds) in which FastQ records will be collected for alignment
ALIGNMENT_WINDOW=20
# how frequently (in wallclock seconds) are statistics updated for dashboard visualizaiton?
STATS_UPDATE_FREQUENCY=30
# IP address of the aligner cluster created by running aligner/provision_species.sh
SPECIES_ALIGNER_CLUSTER_IP=$(gcloud compute forwarding-rules describe bwa-species-forward --global --format="value(IPAddress)")
# IP address of the aligner cluster created by running aligner/provision_resistance_genes.sh
RESISTANCE_GENES_ALIGNER_CLUSTER_IP=$(gcloud compute forwarding-rules describe bwa-resistance-genes --global --format="value(IPAddress)")
# base URL for http services (bwa and kalign)
# value for species, for resistance_genes use 'SERVICES_HOST=http://$RESISTANCE_GENES_ALIGNER_CLUSTER_IP'
SERVICES_HOST=http://$SPECIES_ALIGNER_CLUSTER_IP
# bwa path
BWA_ENDPOINT=/cgi-bin/bwa.cgi
# bwa database name
BWA_DATABASE=DB.fasta
# kalign path
KALIGN_ENDPOINT=/cgi-bin/kalign.cgi
# Firestore DB url defined above
FIRESTORE_URL=https://upwork-nano-stream.firebaseio.com
# Collection name of the Firestore database that will be used for writing output statistic data
FIRESTORE_COLLECTION_STATS=resistance_sequences_statistic
# Collection name of the Firestore database that will be used for writing output Sequences Body data
FIRESTORE_COLLECTION_RESISTANCE_BODIES=resistance_sequences_bodies
# Collection name of the Firestore database that will be used for saving NCBI genome data cache
FIRESTORE_TAXONOMY_CACHE=resistance_gene_cache
# [optional] Only used in resistance_genes mode. Path to fasta file with resistance genes database
RESISTANCE_GENES_FASTA=gs://nanostream-dataflow-demo-data/gene-info/DB_resistant_formatted.fasta
# [optional] Only used in resistance_genes mode. Path to fasta file with resistant genes list
RESISTANCE_GENES_LIST=gs://nanostream-dataflow-demo-data/gene-info/resistance_genes_list.txt
To start Nanostream Pipeline run following command:
java -cp (path_to_nanostream_app_jar) \
com.theappsolutions.nanostream.NanostreamApp \
--runner=$RUNNER \
--project=$PROJECT \
--streaming=true \
--processingMode=$PROCESSING_MODE \
--inputDataSubscription=$UPLOAD_SUBSCRIPTION \
--alignmentWindow=$ALIGNMENT_WINDOW \
--statisticUpdatingDelay=$STATS_UPDATE_FREQUENCY \
--servicesUrl=$SERVICES_HOST \
--bwaEndpoint=$BWA_ENDPOINT \
--bwaDatabase=$BWA_DATABASE \
--kAlignEndpoint=$KALIGN_ENDPOINT \
--outputFirestoreDbUrl=$FIRESTORE_URL \
--outputFirestoreSequencesStatisticCollection=$FIRESTORE_COLLECTION_STATS \
--outputFirestoreSequencesBodiesCollection=$FIRESTORE_COLLECTION_RESISTANCE_BODIES \
--outputFirestoreGeneCacheCollection=$FIRESTORE_TAXONOMY_CACHE \
--resistanceGenesFastDB=$RESISTANCE_GENES_FASTA \
--resistanceGenesList=$RESISTANCE_GENES_LIST
For this project the bucket nanostream-dataflow-demo-data were created with reference databases of species and antibiotic resistance genes.
The bucket has a structure like:
gs://nanostream-dataflow-demo-data/
|- reference-sequences/
|-- antibiotic-resistance-genes/
|--- DB.fasta
|--- DB.fasta.[amb,ann,bwt,pac,sa]
|-- species/
|--- DB.fasta
|--- DB.fasta.[amb,ann,bwt,pac,sa]
where:
- DB.fasta - FASTA file with reference sequences
- DB.fasta.amb, DB.fasta.ann, DB.fasta.bwt, DB.fasta.pac, DB.fasta.sa - files generated and used by
bwain order to improve performance, see details in this SEQanswers answer
nanostream-dataflow-demo-data - is a public bucket with requester pays option enabled.
To build jar from source, follow next steps:
- Install Maven
- Add Japsa 1.9-2b package to local Maven repository. To do this you should run following command from project root:
mvn install:install-file -Dfile=NanostreamDataflowMain/libs/japsa.jar -DgroupId=coin -DartifactId=japsa -Dversion=1.9-2b -Dpackaging=jar
- Build uber-jar file
cd NanostreamDataflowMain
mvn clean package
after running this command successfully, there should be a file: "NanostreamDataflowMain/target/NanostreamDataflowMain-1.0-SNAPSHOT.jar"