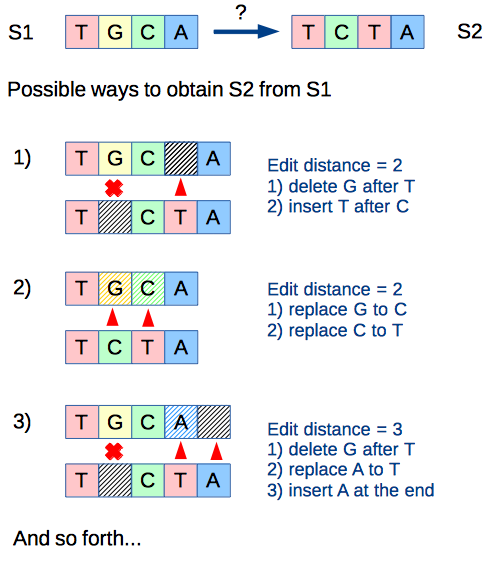
In many applications of the sequence alignment techniques there is an assumption, that the difference between two compared sequences is a result of the minimal amount of the edit operations (thus the calculation of the minimal edit distance is used). However, in some cases there might be a need to analyze the certain amount of the possible sequence alignments (top-K alignments in the order of increase of the edit distance). Thus, the presented algorithm helps to tackle the latter use case.
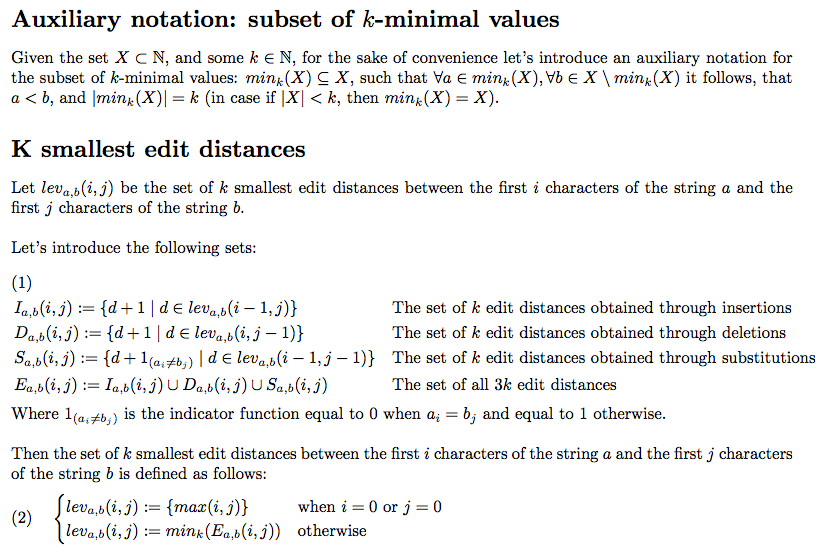
The presented algorithm, finds the top-K different string alignments with the shortest edit distances among other possible alignments (based on the Levenshtein distance definition). Below is a short description of the algorithm, which complies with the terminology of the standard Levenshtein distance algorithm: https://en.wikipedia.org/wiki/Levenshtein_distance
Let's denote M as the length of the input string s1, and N is the length of the input string s2.
Then, the runtime complexity of the algorithm is O(N*M*K + K*log(K)) or O(N*M*K*log(K)) (depending on the implementation of the supplementary procedure for selection of the K smallest elements of an array).
The implementation of the algorithm is in the file: src/main/java/com/lahodiuk/levenshtein/topk/LevenshteinTopK.java
The demonstration of the algorithm is in the file: src/main/java/com/lahodiuk/levenshtein/topk/Demo.java
Example of input:
s1 = "TGCA"
s2 = "TCTA"
K = 10
Example of output:
Total amount of results: 10
=======================================
Edit distance:2
s1 aligned: TGC_A
s2 aligned: T_CTA
common str: T_C_A
=======================================
Edit distance:2
s1 aligned: TGCA
s2 aligned: TCTA
common str: T__A
=======================================
Edit distance:3
s1 aligned: TGCA_
s2 aligned: T_CTA
common str: T_C__
=======================================
Edit distance:3
s1 aligned: TG_CA
s2 aligned: TCT_A
common str: T___A
=======================================
Edit distance:3
s1 aligned: T_GCA
s2 aligned: TCT_A
common str: T___A
=======================================
Edit distance:3
s1 aligned: TGC_A
s2 aligned: TC_TA
common str: T___A
=======================================
Edit distance:3
s1 aligned: TGC_A
s2 aligned: _TCTA
common str: __C_A
=======================================
Edit distance:3
s1 aligned: T_GCA
s2 aligned: TC_TA
common str: T___A
=======================================
Edit distance:3
s1 aligned: TG_CA
s2 aligned: T_CTA
common str: T___A
=======================================
Edit distance:4
s1 aligned: TGC__A
s2 aligned: T_CTA_
common str: T_C___
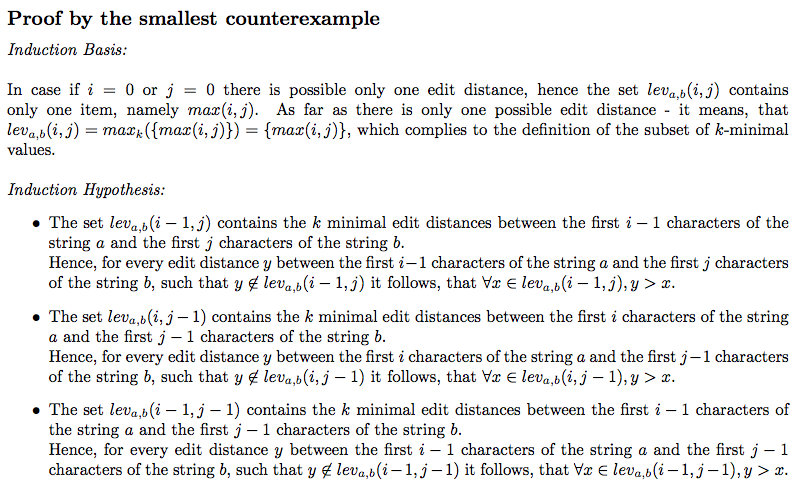
The whole proof is in the levenshtein_top_k.pdf