Please see our Neuroinformatics Paper for more information and cite if using:
Sharrock, M.F., Mould, W.A., Ali, H. et al. 3D Deep Neural Network Segmentation of Intracerebral Hemorrhage: Development and Validation for Clinical Trials. Neuroinformatics (2020). https://doi.org/10.1007/s12021-020-09493-5
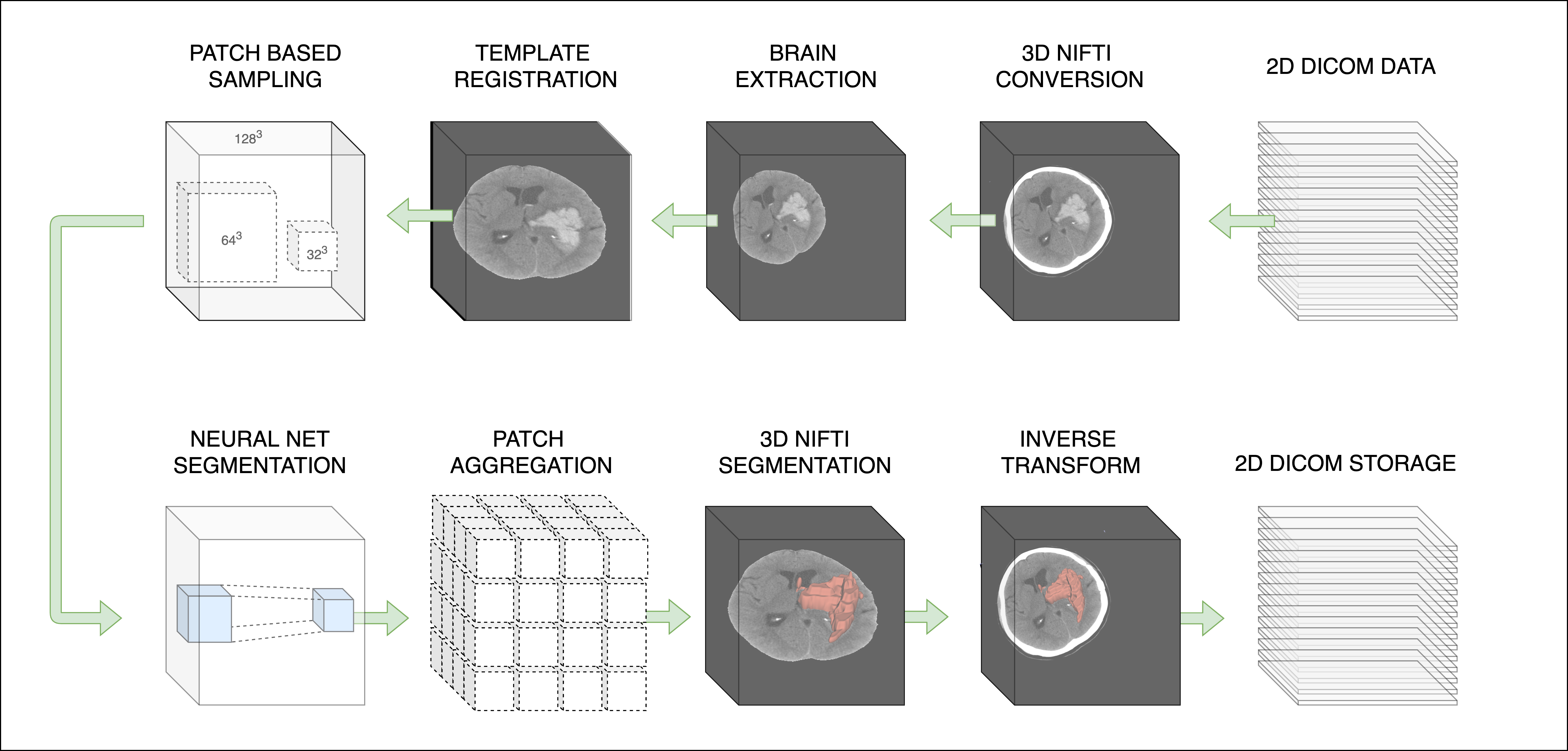
ICH volumetric estimation is a task routinely performed in clinical research. This is the first publicly available deep neural network model to perform the task of ICH segmentation and volume estimation. We showed in our paper that 3D segmentation with appropriate anatomic context presented in the receptive field outperforms 2D segmentation and state-of-the-art results can be obtained on smaller datasets with standard publicly available models. This model expects non-contrast CT in NIfTI format and automatically performs preprocessing including a validated CT brain extraction method and spatial normalization via registration to a 1.5mm x 1.5mm x 1.5mm CT template with a shape of (128, 128, 128).
To run an ICH prediction set the path to directories for inputs, outputs and model weights:
$ python3 predict.py --indir /path/to/inputs/ --outdir /path/to/outputs/ --weights /path/to/weights | Train Dataset | Val Dataset | Bleeds | Val Dice | Weights Link |
|---|---|---|---|---|
| MISTIE 2 | MISTIE 3 | ICH, IVH | 0.92 | m2_weights |
On some systems you may need to rename the files beginning with a . to a common prefix, such as weights (e.g. .index becomes weights.index).
Here is a script to automatically download them:
wget -O weights.zip https://www.dropbox.com/s/v2ptd9mfpo13gcb/mistie_2-20200122T175000Z-001.zip?dl=1
unzip -j weights.zip
for i in _data-00001-of-00002 _data-00000-of-00002 _index;
do
out=`echo ${i} | sed "s/_/weights./"`;
mv ${i} ${out};
doneExample Data Set
To see how the model performs, you can download data from a head CT brain extraction validation paper: https://archive.data.jhu.edu/dataset.xhtml?persistentId=doi:10.7281/T1/CZDPSX
For example, here is how to download and extract an image in a directory called input:
wget --no-check-certificate https://archive.data.jhu.edu/api/access/datafile/1311?gbrecs=true -O 01.tar.xz
tar xvf 01.tar.xz
mkdir -p input
mv 01/BRAIN_1_Anonymized.nii.gz input/image.nii.gzRunning the command, assuming the weights were downloaded and extracted as above and the input directory is input as done above, would be the command:
python3 predict.py --verbose --indir ./input --outdir ./output --weights weightsand the resulting mask would be in the output directory.
| Option | Argument | Example |
|---|---|---|
| Number GPUs | --gpus | --gpus 2 |
| Number CPUs | --cpus | --cpus 8 |
| Verbose + Timing | --verbose | --verbose |
Setup
Current dependencies are listed below.
| Software | Version | URL |
|---|---|---|
| Tensorflow | 2.1.0 | https://www.tensorflow.org |
| ANTsPy | 0.1.7 | https://github.com/ANTsX/ANTsPy |
| FSLPy | 2.7.0 | https://git.fmrib.ox.ac.uk/fsl/fslpy |
| FSL* | 6.0.2 | https://fsl.fmrib.ox.ac.uk/fsl/fslwiki |
*Please read and agree to the software license for FSL prior to use.
DeepBleed Docker
Alternatively, you can pull a pre-built docker image with the dependencies installed:
$ docker pull msharrock/deepbleed To run a prediction, start the docker image, link the data path that contains the indir and outdir:
$ docker run -it msharrock/deepbleed bash -v /path/to/data:/data/ Pull this repository from github and then run predictions as previously noted
Directions for OSX
For OSX, you should likely install ANTsPy using a git install of:
pip install git+git://github.com/ANTsX/ANTsPy.gitBut you can try a release from the releases page: https://github.com/ANTsX/ANTsPy/releases
python3.7 -m pip install https://github.com/ANTsX/ANTsPy/releases/download/v0.1.8/antspyx-0.1.8-cp37-cp37m-macosx_10_14_x86_64.whlLicense
Please see LICENSE.md for information of the license and uses.