ddtlcm: Dirichlet diffusion tree-latent class model (DDT-LCM)
An R package for Tree-regularized latent class mModels with a DDT process prior on class profiles
Maintainer: Mengbing Li (mengbing@umich.edu)
Contributors: Briana Stephenson (bstephenson@hsph.harvard.edu); Zhenke Wu (zhenkewu@umich.edu)
| Citation | Paper Link | |
|---|---|---|
| Bayesian tree-regularized LCM | Li M, Stephenson B, Wu Z (2023). Tree-Regularized Bayesian Latent Class Analysis for Improving Weakly Separated Dietary Pattern Subtyping in Small-Sized Subpopulations. ArXiv:2306.04700. | Link |
# install bioconductor package `ggtree` for visualizing results:
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("ggtree")
install.packages("devtools",repos="https://cloud.r-project.org")
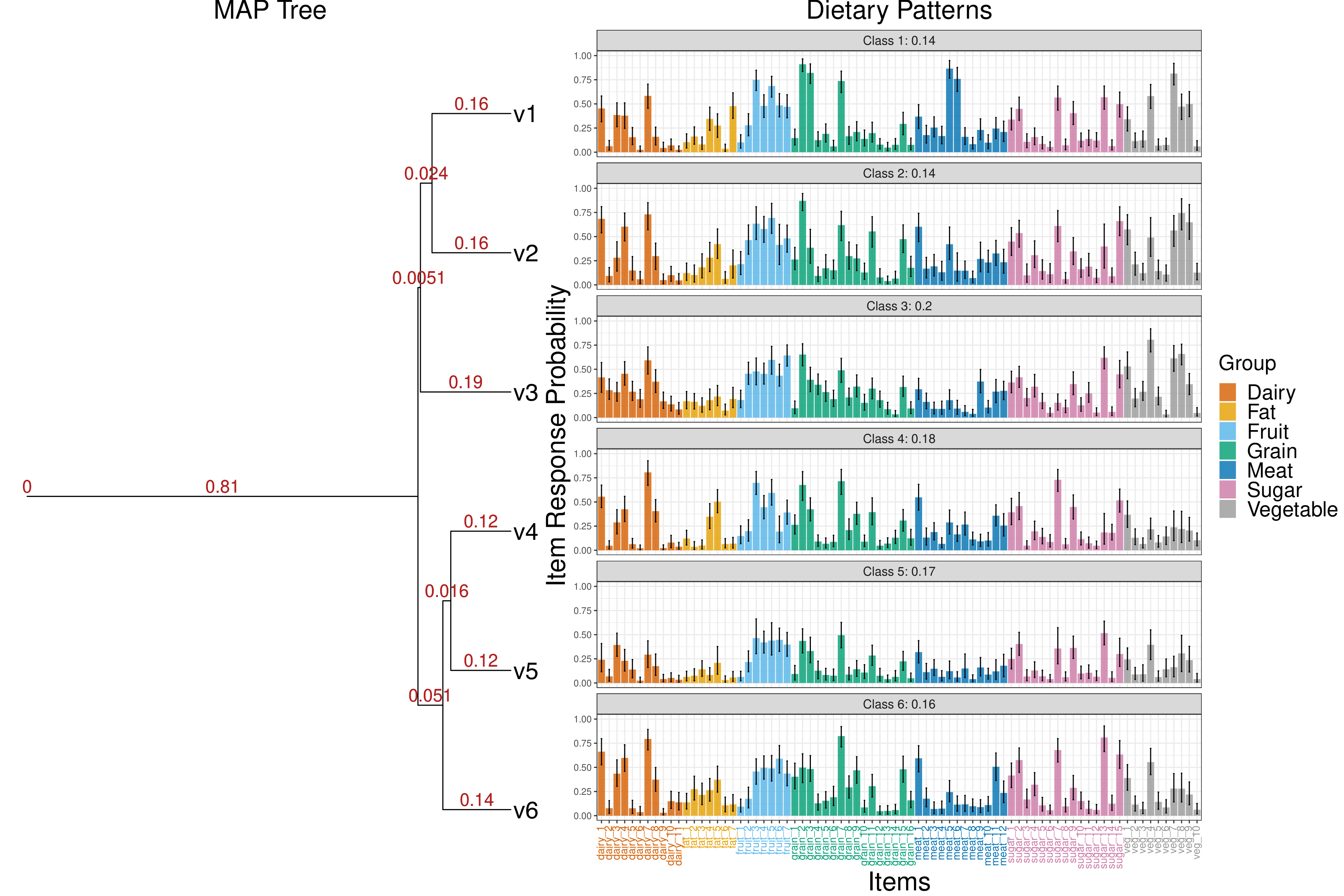
devtools::install_github("limengbinggz/ddtlcm")ddtlcm is designed for analyzing multivariate binary observations over grouped items in a tree-regularized Bayesian LCM framework. Between-class similarities are guided by an unknown tree, where classes positioned closer on the tree are more similar a priori. This framework facilitates the sharing of information between classes to make better estimates of parameters using less data. The model is built upon equipping LCMs with a DDT process prior on the class profiles, with varying degrees of shrinkage across major item groups. The model is particularly promising for addressing weak separation of latent classes when sample sizes are small. The posterior inferential algorithm is based on a hybrid Metropolis-Hastings-within-Gibbs algorithm and can provide posterior uncertainty quantifications.
ddtlcm works for
-
multivariate binary responses over pre-specified grouping of items
-
The functions' relations in the package
ddtlcmcan be visualized by
library(DependenciesGraphs) # if not installed, try this-- devtools::install_github("datastorm-open/DependenciesGraphs")
library(QualtricsTools) # devtools::install_github("emmamorgan-tufts/QualtricsTools")
dep <- funDependencies('package:ddtlcm','ddtlcm_fit')
plot(dep)