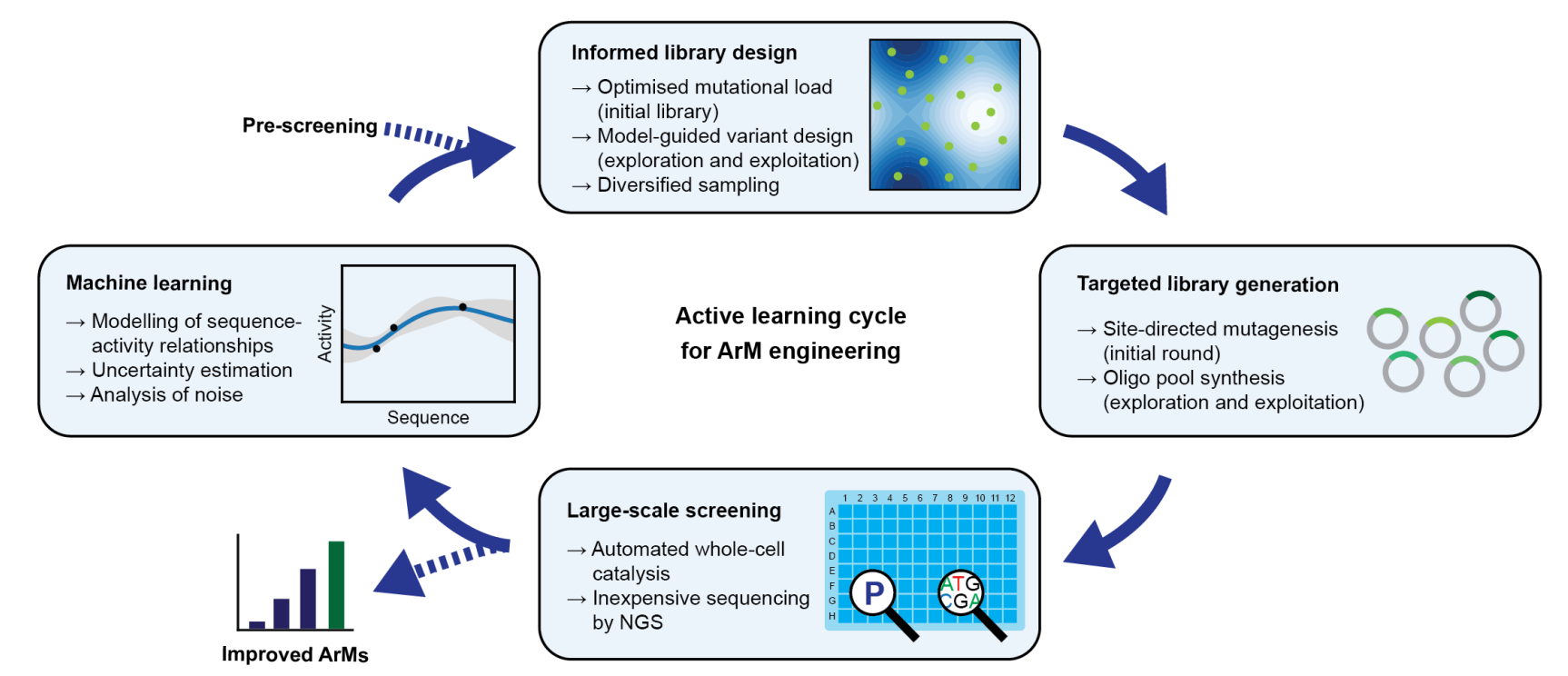
Content: Enhanced Sequence-Activity Mapping and Evolution of Artificial Metalloenzymes by Active Learning
This repository contains scripts for analysis, preparation and reporting of results from a study published in ACS Central Science journal:
Authors: Tobias Vornholt Mojmír Mutny, Gregor W. Schmidt, Christian Schellhaas, Ryo Tachibana, Sven Panke, Thomas R. Ward, Andreas Krause, and Markus Jeschek
Title: Enhanced Sequence-Activity Mapping and Evolution of Artificial Metalloenzymes by Active Learning
Journal: ACS Central Science
Year: 2024
For the full-paper please refer to the link.
This repository containts:
- plotting scripts in
/plots - training scripts in
bechmark_run - sequential decision making scripts in
active_learning_X
As part of the project we have developed a standalone python package mutedpy, which can be found in the dependecies section.
- 21/05/2024 Initial version of public code online
This repository contains only basic script which build upon libraries
![]() 1.
1. mutedpy https://github.com/Mojusko/mutedpy
 2.
2. stpy https://github.com/Mojusko/stpy
A large part of the dataset could not fit to the repository. Additional data is located in
- Embeddings form the
/datacan be downloaded from here. - NGS sequencing analysis can be downloaded from here.
- 10% subset of structured generated via Rosetta software. It can be downloaded here.
- Pretrained and saved models for the plotting can be found here.
The easiest way to rerun the clode is to clone repository along with the stpy repository as
git clone https://github.com/Mojusko/mutedpy
cp /experiment/
git clone https://github.com/lasgroup/ml-protein-design-sav-gold
mv ml-protein-design-sav-gold streptavidin
cd streptavidin
wget https://polybox.ethz.ch/index.php/s/XKNUFIGRY08py63 #retrieve saved embeddings data
unzip data.zip data
wget https://polybox.ethz.ch/index.php/s/Bd9bi0ITfBI6xur #retrieve save pickled models
uzip models.zip modelsThe benchmarking analysis can be rerun using the code in bechmark_run sub-folder. Namely, the final parameters for the chemical features can be run with:
cd experiments/streptavidin
mkdir results_strep
cd bechmark_run
mkdir job_files_exp
python benchmark_run/run_final_aa.py
sh job_files_exp/job0.shThe final model is then saved to to the results_strep subfolder along with plots of different cross-validation splits. To rerun the extensive benchmark access to our MongoDB database is needed. The code and calculation of structure is however available online. We used benchmark_run/run_extra_analysis_bench_2.py to generate hyperparameter for benchmarking.
The plots for the publication and statistical analysis can be found in the subfolder plots/.
To cite this work, please use
@article{Vornholt2024,
author = {Vornholt*, Tobias and Mutn{\'y}*, Mojm{\'\i}r and Schmidt, Gregor and Schellhaas, Christian and Tachibana, Ryo and Panke, Sven and Ward, Thomas R. and Krause, Andreas and Jeschek, Markus},
journal = {ACS Central Science},
title = {{Enhanced Sequence-Activity Mapping and Evolution of Artificial Metalloenzymes by Active Learning}},
url = {https://www.biorxiv.org/content/10.1101/2024.02.06.579157v1.full.pdf},
year = {2024}
}
This repository was assembled by Mojmir Mutny (ETH Zuerich) and Tobias Vornholt (ETH Zuerich and University of Basel).
For any inquries regarding the code, please use: mmutny@inf.ethz.ch