Overlap-layout-consensus based DNA assembler of long uncorrected reads (short for Rapid Assembler).
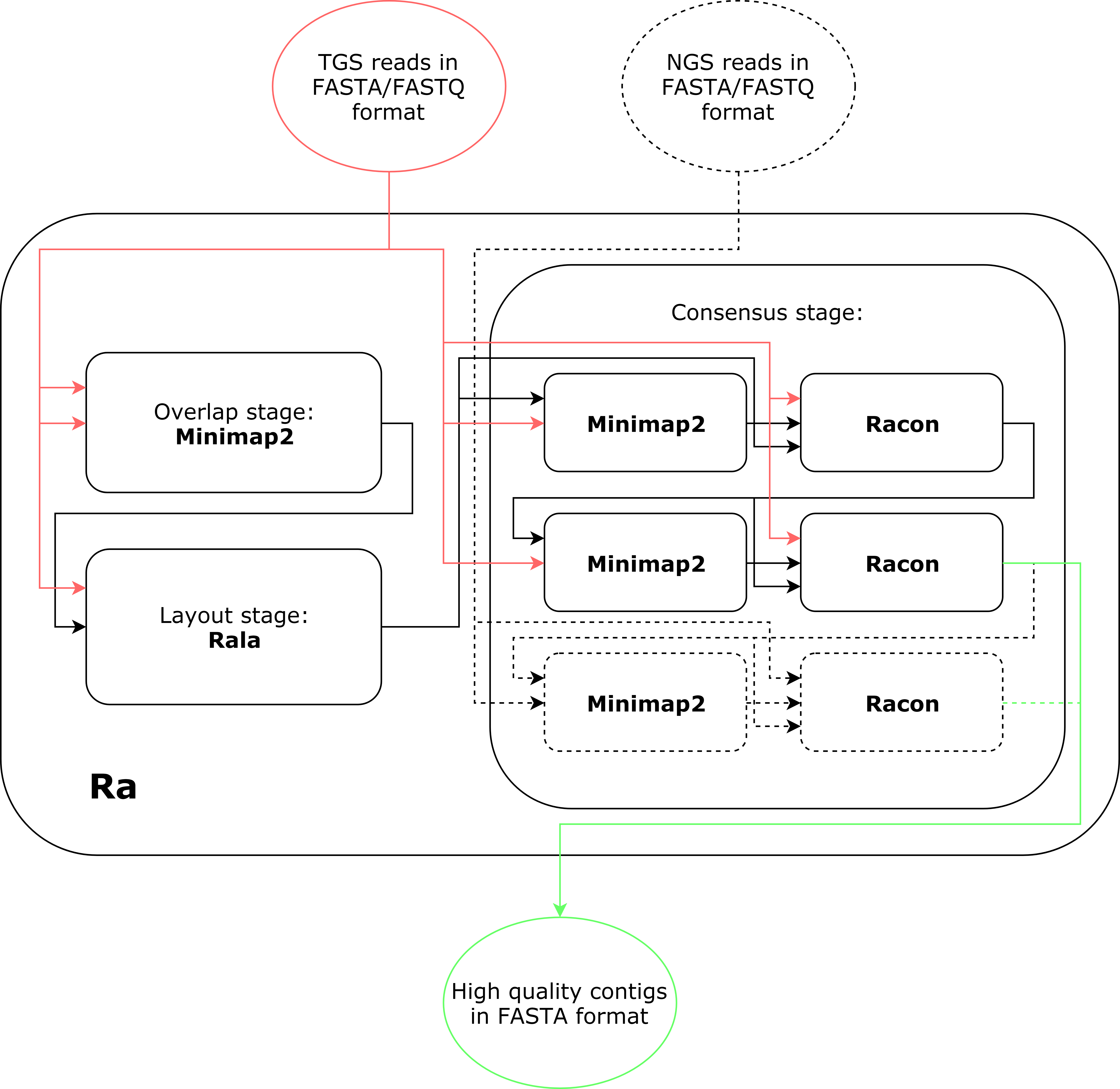
Ra is as a fast and easy to use assembler for raw reads generated by third generation sequencing. It consists of Minimap2, Rala and Racon as depicted on figure bellow.
Ra takes as input a single file containing raw reads in FASTA/FASTQ format (can be compressed with gzip) and outputs a set of contigs with high accuracy in FASTA format to stdout. Additionally, a file containing second generation sequences in FASTA/FASTQ format (can be compressed with gzip) can be passed as second argument in order to polish the final assembly.
- gcc 4.8+ or clang 3.4+
- cmake 3.2+
To install Ra run to following commands:
git clone --recursive https://github.com/rvaser/ra.git ra
cd ra
mkdir build
cd build
cmake -DCMAKE_BUILD_TYPE=Release ..
makeAfter successful installation, an executable named ra will appear in build/bin.
Note: if you omitted --recursive from git clone, run git submodule update --init --recursive before proceeding with compilation.
Usage of ra is as following:
ra [options ...] -x {ont, pb} <sequences> [<ngs_sequences>]
<sequences>
input file in FASTA/FASTQ format (can be compressed with gzip)
containing third generation sequencing reads
<ngs_sequences>
input file in FASTA/FASTQ format (can be compressed with gzip)
containing next generation sequencing reads
required arguments:
-x {ont, pb}
sequencing technology of input sequences
options:
-t, --threads <int>
default: 1
number of threads
--version
prints the version number
-h, --help
prints the usage
For additional information, help and bug reports please send an email to: robert.vaser@fer.hr.
This work has been supported in part by Croatian Science Foundation under the project UIP-11-2013-7353.