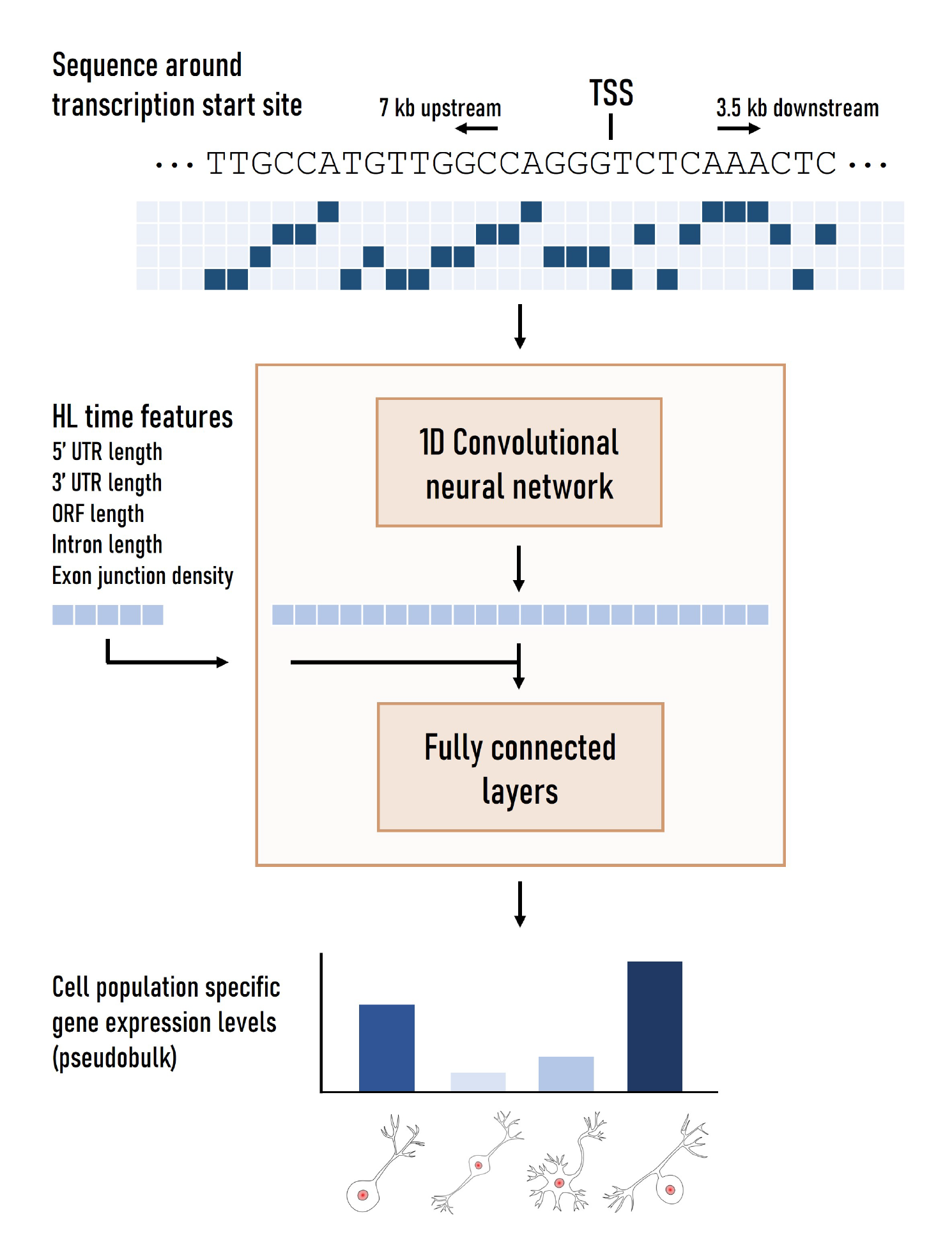
We present scXpresso: a method that predicts gene expression at an unprecedented resolution using single-cell RNA-sequencing data. Using this GitHub, you can reproduce the figures from the paper, train your own models, or look at the predicted effect of interesting variants.
The results, trained models, input sequences etc. can all be downloaded from Zenodo.
This folder contains the code to train your own models. Python version 3.6 or higher and PyTorch version 1.9 or higher is required for this.
Here, we explain step-by-step how to train your own models and look at interesting variants or apply in-silico mutagenesis.
- Creating your own pseudobulk values
- Training scXpresso models
- Evaluating the performance
- In-silico mutagenesis
- Looking at individual variants
Contains Jupyter Notebooks to reproduce all the figures from the preprint.
For citation and further information please refer to: "Predicting cell population-specific gene expression from genomic sequence"