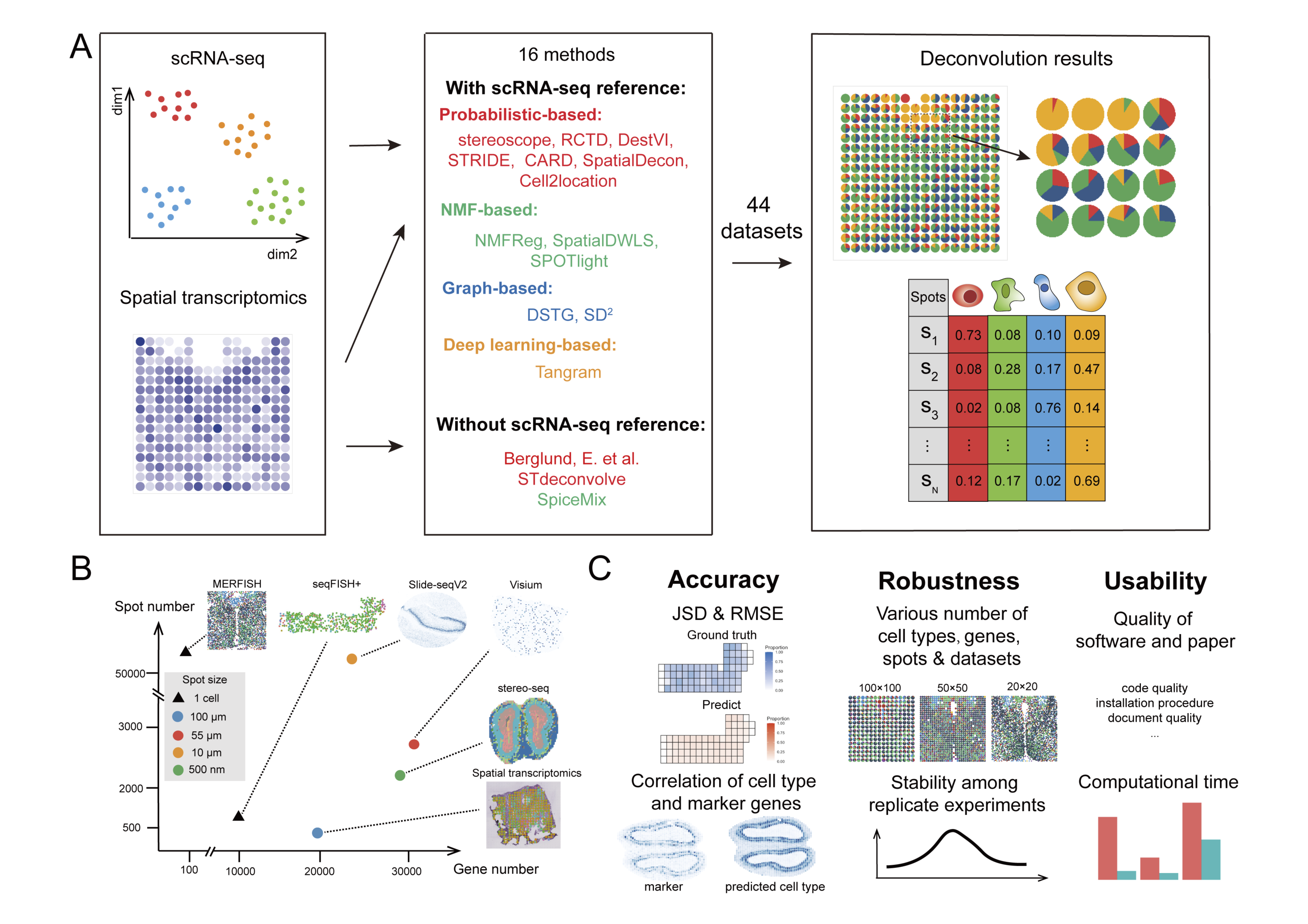
We benchmarked 16 existing methods resolving a cellular deconvolution task with 44 real-world and simulated datasets by evaluating the accuracy, robustness, and usability of the methods. For accuracy, We used JSD score, RMSE and Pearson correlation coefficient to quantify the performance of all methods on the datasets from six image-based and sequencing-based spatial transcriptomics technologies. For robustness, we designed several experiments with different conditions as follows: (1) 10,000, 6000, or 3000 genes were randomly chosen in the seqFISH+ dataset; (2) three resolutions were simulated with 12 MERFISH datasets using binning sizes of 20, 50, and 100 μm, which were also used to test performance with different numbers of spots; (3) 17 original cells types and 11 integrated cell types were tested in Slide-seqV2 datasets; and (4) the stability of the performance was assessed by repeating the experiments three times with the seqFISH+ dataset using 10,000 genes per spot. For usability, we recorded the running time with three different spot numbers in the MERFISH datasets and scored several aspects of the tutorials of all methods, including document quality, code quality, installation procedure, and example analysis.
For implementation, we followed the codes from all proposed methods with their default parameters. In the /methods, we showed the implementation of all 16 methods on seqFISH+ dataset as an example. Please make sure the installation of all methods following these versions: CARD (v1.0.0),SPOTlight (v0.99.0), DSTG, SpatialDWLS, SD2, NMFreg,
stereoscope (v.03), Tangram (v1.0.3), Cell2location (v0.1), STdeconvolve (1.0.0), Berglund (0.2.0), SpiceMix, RCTD (spacexr 2.0.0), STRIDE, DestVI (scvi-tools 0.16.0) and SpatialDecon.
In the /evaluation & visualization, we implemented the visualization and quantification of all figures in our manuscript and supplementary materials.
To test all methods on example dataset (seqFISH+ with 10000 genes), you could download the file and put it into /methods.
Each method contains a file including the README file and its implementation on example dataset.