This repository contains an end-to-end pipeline to reproduce and extend the dataset curation, data shift quantification and empricial evaluation presented in the paper:
Drug Discovery under Covariate Shift with Domain-Informed Prior Distributions over Functions. Leo Klarner, Tim G.J. Rudner, Michael Reutlinger, Torsten Schindler, Garrett M. Morris, Charlotte M. Deane, Yee Whye Teh ICML 2023.
— View Paper —
Abstract: Accelerating the discovery of novel and more effective therapeutics is an important pharmaceutical problem in which deep learning is playing an increasingly significant role. However, real-world drug discovery tasks are often characterized by a scarcity of labeled data and significant covariate shift—a setting that poses a challenge to standard deep learning methods.
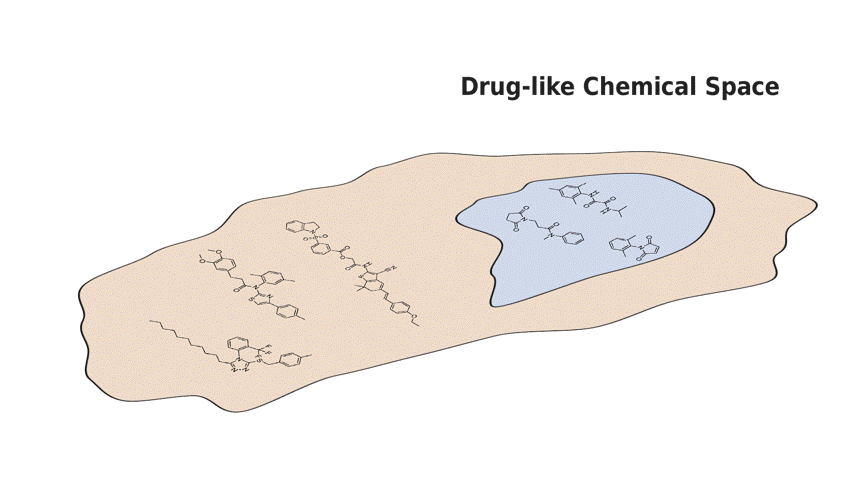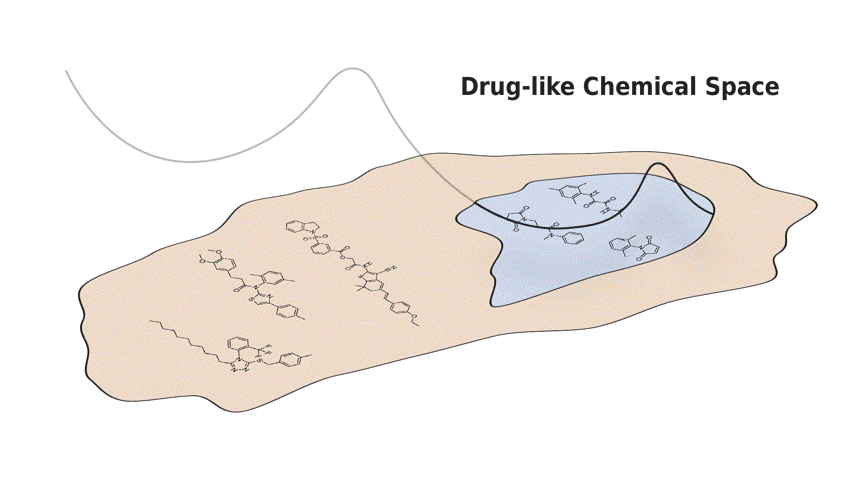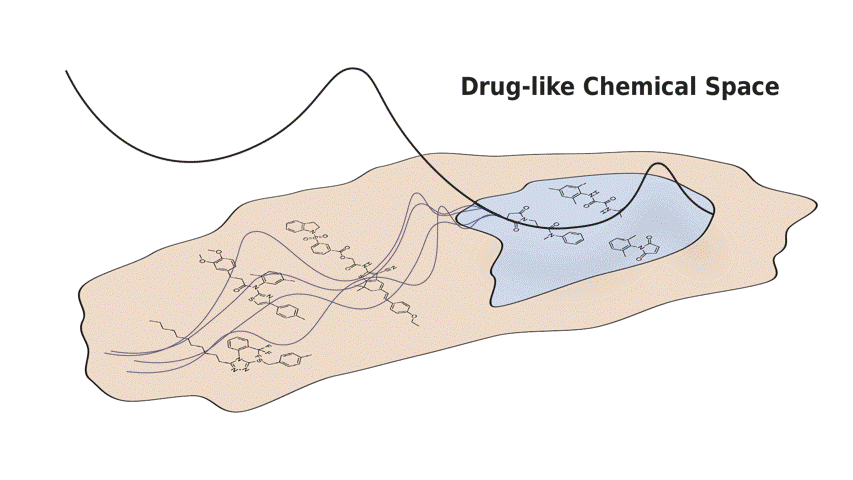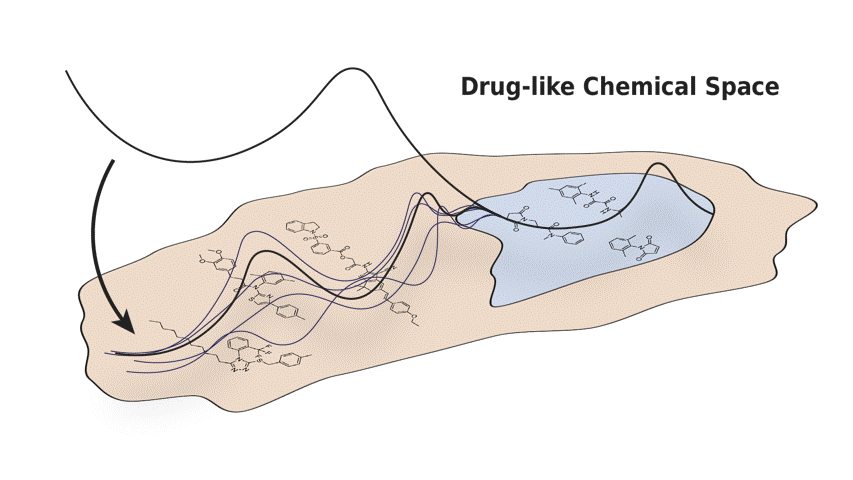 In this paper, we present Q-SAVI, a probabilistic model able to address these challenges by encoding explicit prior knowledge of the data-generating process into a prior distribution over functions, presenting researchers with a transparent and probabilistically principled way to encode data-driven modeling preferences. Building on a novel, gold-standard bioactivity dataset that facilitates a meaningful comparison of models in an extrapolative regime, we explore different approaches to induce data shift and construct a challenging evaluation setup. We then demonstrate that using Q-SAVI to integrate contextualized prior knowledge of drug-like chemical space into the modeling process affords substantial gains in predictive accuracy and calibration, outperforming a broad range of state-of-the-art self-supervised pre-training and domain adaptation techniques.
In this paper, we present Q-SAVI, a probabilistic model able to address these challenges by encoding explicit prior knowledge of the data-generating process into a prior distribution over functions, presenting researchers with a transparent and probabilistically principled way to encode data-driven modeling preferences. Building on a novel, gold-standard bioactivity dataset that facilitates a meaningful comparison of models in an extrapolative regime, we explore different approaches to induce data shift and construct a challenging evaluation setup. We then demonstrate that using Q-SAVI to integrate contextualized prior knowledge of drug-like chemical space into the modeling process affords substantial gains in predictive accuracy and calibration, outperforming a broad range of state-of-the-art self-supervised pre-training and domain adaptation techniques.
All Q-SAVI models and objectives are implemented in JAX/Haiku. The repository is structured as follows:
data/contains the both the raw and processed data, as well as all processing utilities required to derive the anti-maralarial dataset and the ZINC-based context point distribution.datasets/contains the raw and processed anti-malarial dataset, as well as ~2m unlabeled molecular structures from the ZINC database.preprocess_antimalarial_data.ipynbannotated notebook that describes all procedures used for data curation, covariate and label shift quantification, and data splitting.preprocess_zinc.pyutilities to convert ZINC SMILES strings to ECFPs and rdkitFPs.
qsavi/contains all models, objectives and utilities needed to reproduce and extend the results presented in the paper.bayesian_mlps.pydefinition of stochastic MLPs used in the paper.config.pydefault hyperparameter settings and search spaces.context_points.pyfunctions to sample from pre-processed context point distribution.data_loader.pydata loading and processing utilities.linearization.pylinearization utilities for the objective evaluation.objective.pyimplementation of the function-space objective presented in the paper.qsavi.pyQ-SAVI class that combines stochastic MLPs with function-space objective.utils.pymiscellaneous utilities.
The easiest way to get started with Q-SAVI is to run the provided Colab notebook. It takes care of the installation and setup for you, and provides a step-by-step demonstration on how to train and evaluate Q-SAVI models and reproduce the experimental results presented in the paper.
Download the Q-SAVI source code from GitHub and set up a virtual environment with the appropriate JAX version (see https://github.com/google/jax#installation for more details).
git clone -b staging https://github.com/leojklarner/Q-SAVI.git
cd Q-SAVI
python -m venv qsavi_env
source qsavi_env/bin/activate
python -m pip install --upgrade pip
python -m pip install --upgrade jax==0.4.7
python -m pip install https://storage.googleapis.com/jax-releases/cuda11/jaxlib-0.4.7+cuda11.cudnn82-cp310-cp310-manylinux2014_x86_64.whl
python -m pip install --upgrade -r requirements.txt
Download and extract the pre-processed context point distribution, i.e. a uniform subsample of the ZINC database featurized as extended connectivity fingerprints.
mkdir data/datasets/zinc
wget https://www.dropbox.com/s/xsbz8wyewupnpe8/zinc_context_points_ecfp.tar.gz?dl=0 -P data/datasets/zinc
tar -xf data/datasets/zinc/zinc_context_points_ecfp.tar.gz?dl=0 -C data/datasets/zinc
The Q-SAVI models are instantiated as a qsavi.QSAVI class, which combines a stochastic MLP with the function-space objective. All hyperparameter options are specified in qsavi/config.py. The following code snippet demonstrates how to instantiate a Q-SAVI model with the default hyperparameter settings.
import argparse
from qsavi.config import add_qsavi_args
from qsavi.qsavi import QSAVI
def run_qsavi():
parser = argparse.ArgumentParser()
add_qsavi_args(parser)
kwargs = parser.parse_args()
kwargs.split = "spectral_split"
kwargs.featurization = "ec_bit_fp"
kwargs.learning_rate = 1e-4
kwargs.num_layers = 2
kwargs.embed_dim = 32
kwargs.prior_cov = 100.0
kwargs.n_context_points = 16
kwargs.datadir = "data/datasets"
qsavi = QSAVI(kwargs)
val_metrics, test_metrics = qsavi.train()
return val_metrics, test_metrics
The Q-SAVI model can be applied to new datasets by implementing a custom data loader, see qsavi/data_loader.py. Similarly, a customized prior distribution over the induced function space of the MLP can be specified by adapting the prior function in qsavi/objective.py.
If you found our paper or code useful, please cite it as:
@InProceedings{klarner2023qsavi,
title = {Drug Discovery under Covariate Shift with Domain-Informed Prior Distributions over Functions},
author = {Klarner, Leo and Rudner, Tim G. J. and Reutlinger, Michael and Schindler, Torsten and Morris, Garrett M and Deane, Charlotte and Teh, Yee Whye},
booktitle = {Proceedings of the 40th International Conference on Machine Learning},
pages = {17176--17197},
year = {2023},
volume = {202},
series = {Proceedings of Machine Learning Research},
publisher = {PMLR},
}
