This repository is a template you can use to create new projects.
IMPORTANT: If you are going to work on an existing project, do not clone this repository. You need to look for the repository for that project.
Install GIT Windows
Install GIT LFS
Install Anaconda
Install Github Desktop, if you do not like to use the terminal so much.
Install Visual Studio Build Tools
Install Atom
This can either be done with Github Desktop
or with a terminal. When we say terminal, in Windows we mean Anaconda prompt.
However, all code that starts with git can be done with Github Desktop.
Make a folder where you want to have it, e.g. c:\apps\
With the terminal run
cd c:\apps
git clone https://github.com/CINPLA/cinpla-base.git
Windows: Search for anaconda and open Anaconda prompt.
Mac: open a terminal
Create a new Anaconda environment with (replace myproject with the name of your project):
conda create -n myproject python=3.6
Then, enter the environment using
activate myproject
Navigate to where you have cloned cinpla-base, then install the cinpla-base
requirements
cd cinpla-base
pip install -r requirements.txt
In order to add lab-specific expipe commands, run:
expipe config global -a plugin expipe_plugin_cinpla
In order to run spike sorting with the machine you are using, you have to install them.
Navigate to where you have cloned cinpla-base, then install the cinpla-base
requirements-spiketools. Choose the appropriate file (windows-linux)
cd cinpla-base
pip install -r requirements-spiketools-windows.txt (or requirements-spiketools-linux.txt)
This will install the Python-based sorters(klusta, spyking-circus, mountainsort, herding spikes, tridesclous)
Kilosort2, Ironclust, and WaveClus are matlab-based spike sorters. To install them navigate to C:\apps and run:
git clone https://github.com/MouseLand/kilosort2.git
git clone https://github.com/jamesjun/ironclust.git
git clone https://github.com/csn-le/wave_clus.git
In order to let the system know where these packages are installed we have to set environment variables.
If you have admin access, in Windows, select start-->Computer-->right-click-->Properties.
Then click on Advanced settings-->Environment variables and add this three New variables:
name: KILOSORT2_PATH variable: C:\apps\kilosort2
name: IRONCLUST_PATH variable: C:\apps\ironclust
name: WAVECLUS_PATH variable: C:\apps\wave_clus
In case you don't have admin access you can set temporary environment variables from the anaconda prompt by running:
set KILOSORT2_PATH=C:\apps\kilosort2
set IRONCLUST_PATH=C:\apps\ironclust
set WAVECLUS_PATH=C:\apps\wave_clus
Navigate to where you have cloned cinpla-base, then install the cinpla-base
requirements-curationtools
cd cinpla-base
pip install -r requirements-curationtools.txt
Next you can install phy:
pip install --pre --upgrade phy
(NOTE: On Windows you need an extra package: pip install pywin32)
To add a remote server for spike sorting, run:
expipe add-server -n name-of-the-server(torkel-beist) -d IP-address -un username
You will be prompted a password for the server.
- Contact Mikkel, Alessio or Svenn-Arne
- Navigate to Gitea and expipe on NIRD
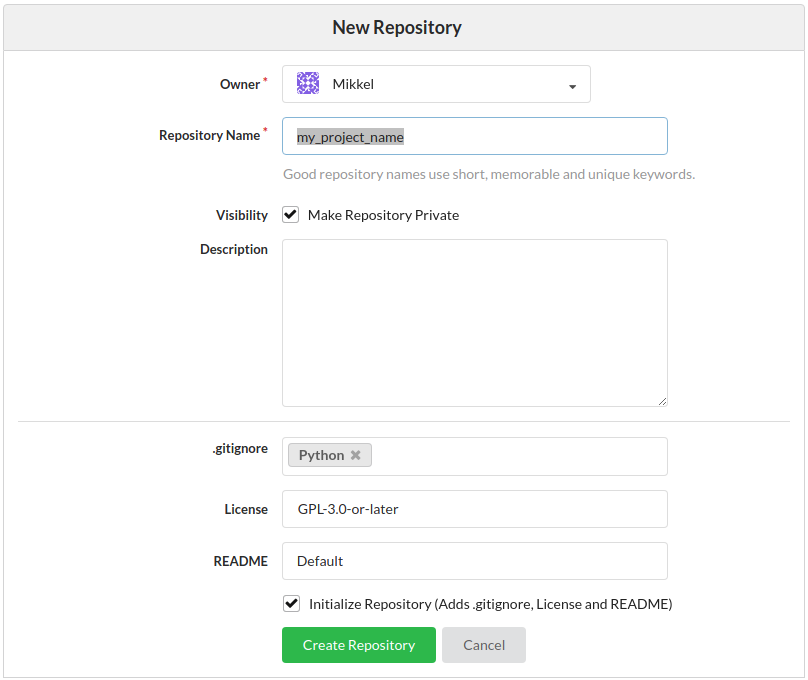
- Initialize with .gitignore, licence and README
- from notebook (see example below)
- with git desktop
If you want to clone a LFS repository and only get the pointer files (not the large files) do
git lfs install --skip-smudge
And then clone as usual
git clone https://gitea.expipe.sigma2.no/user_name/my_project_name.git
Navigate to my_project_name and write in the terminal
jupyter notebook
In stead of using the terminal from now on, commands can be run from within the
notebook if it is begun with an exclamation mark. That is, if you would write
expipe init in the terminal, you would write in the notebook
expipe init
This command adds actions, entities and templates folders together with an
expipe.yaml, this is necesarry so that my_project_name will be recognized as
a expipe project.
Intro from atlassian Official docs
There are many good tutorials for LFS, use those or look at the brief intro below.
Normal git is not "wired" to handle large files, this is why we use git LFS which is helping us handling
large files (LFS stands for Large File Storage)
Be sure to initialize LFS properly before adding and committing files. It is possible to rewrite history later (with git lfs migrate import), but this can be a pain..
git lfs install
Next we need to tell LFS which files should be tracked as large files, if you are not sure, you can add all data which is put in the action/data folder by
git lfs track "actions/*/data/**/*"
Open the .gitattributes files and make sure it reads: (you may want to add the last line which tells LFS to skip .yaml files)
actions/*/data/**/* filter=lfs diff=lfs merge=lfs -text
*.yaml !filter !diff !merge
The first line says that all the contents in all data directories whithin every
action should be handeled by git LFS. The second line says that even though all
the data directories should be handeled by LFS, all files ending with .yaml
should not be handeled by LFS.
In short, these lines ensures that all file except .yaml files will be downloaded
as LFS files when the repository is cloned or pulled if nothing else is specified.
This means that all files in data except .yaml files will be text files
pointing to the real data files on NIRD.
If you only want to track npy and dat files
git lfs track "*.npy"
git lfs track "*.dat"
Add the .gitattributes file
git add .gitattributes
To avoid downloading the original files when doing a git pull the following command adds .lfsconfig
with a line specifying to exclude all LFS tracked files.
git config -f .lfsconfig lfs.fetchexclude "*"
git add .lfsconfig
git commit -am "init expipe and LFS"
Now try to add and commit some files which should be tracked by LFS, and make sure they are correct by looking at
git lfs ls-files
Navigate to cinpla-base/src/expipe-templates-cinpla/templates, where you'll
find a bunch of example templates, copy some you want to use or write your own.
If you want to write your own template the filename must on the form
filename.yaml and at a minimum contain
name: filename
identifier: filename
It is probably wise to commit after you add the templates.
git add -A
git commit -am "added some templates"
And now push your changes:
git push
Now you are ready to start using expipe
from expipe_plugin_cinpla.widgets import browser
browser.display('workshop')
You can register actions (surgery, adjustments, recordings, perfusions, etc.) and process data using the jupyter GUI.
Once you have worked on the repo, you should git add the changed files, git commit and git push.
If you want to physically download the files of an action to perform analysis, for example, first you need to pull the required files:
git pull
git lfs fetch -I path-to-action
git lfs checkout
or equivalently
git -c lfs.fetchexclude="" lfs pull -I path-to-action
If you want to download all files:
git lfs fetch --all
git lfs checkout
Once you have pushed everything, if you want to free some space you can delete your actions and run (do it with caution!!!):
git reset --hard
You can check the status of your expipe project using the expipe Browser:
import expipe
expipe.Browser('workshop').display()
If you want to store your credentials do
git config credential.helper store
git pull
Type in your credentials and it is then stored.
If you get lfs-timeout errors when pushing (i/o timeout, error: failed to push some refs), consider changing your lfs settings to with
This might be a sign that LFS is not tracking your files properly, if so try git lfs migrate import, although we have experienced this not to properly add files to LFS, see which files are tracked by git lfs ls-files
git config lfs.tlstimeout 300
git config lfs.activitytimeout 60
git config lfs.dialtimeout 600
git config lfs.concurrenttransfers 1
git config --global https.postBuffer 2097152000
git config --global http.postBuffer 2097152000
Sometimes when cloning, pulling, or pushing to gitea you might get this error:
fatal: unable to access 'https://gitea.expipe.sigma2.no/username/project.git/': The requested URL returned error: 403
This can be solved by changing the repo name copied from gitea:
https://gitea.expipe.sigma2.no/username/project.git/
to
https://username@gitea.expipe.sigma2.no/username/project.git/
If your .git folder gets huge, you can delete old LFS files from local storage with (make sure you are sync with remote (all files are pushed))
git lfs prune
- if you get a
multiarray errorwhen runningexpipe initrun:
pip uninstall numpy
pip install numpy
No module named repository
pip install gitpython
Stale connection, unable to connect to kernel
pip install "tornado<6"